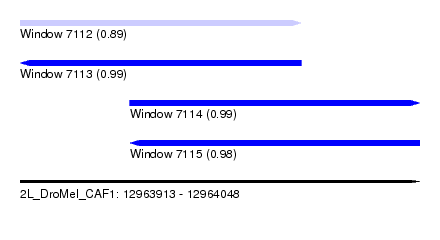
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,963,913 – 12,964,048 |
| Length | 135 |
| Max. P | 0.985768 |

| Location | 12,963,913 – 12,964,008 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12963913 95 + 22407834 GCGAACCUUU--UUUGAUUAUGAUUGGGACACAUGGU-GAAAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAUGGCCAGCAGGUCCU ..........--.............(((((.(((...-...)))---.(((-----------------(((((((.(((....((((.....)))).--..))).))))))))))))))) ( -35.50) >DroPse_CAF1 43482 120 + 1 GCGGACCUUUUUUUUGAUUAUGAUUCGAACACAUGGGCGACACAGGGUUUGGGGGGUAGGGGAGGGGAGGGGCUGACUCAAUAAGGCUAAAAGCCAGCGACGAGGACGCCACGAGGUCCU ..(((((((((((((..((((..(((((((...(((....).))..)))))))..))))..)))))))..(((...(((.....(((.....)))......)))...)))...)))))). ( -35.50) >DroSec_CAF1 8706 95 + 1 GCGAACCUUU--UUUGAUUAUGAUUGGGACACAUGGU-GAAAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGUUGGCCAGCAGGUCCU ..........--.............(((((.(((...-...)))---.(((-----------------(((((((((((....((((.....)))).--..))))))))))))))))))) ( -39.70) >DroSim_CAF1 9522 95 + 1 GCGAACCUUU--UUUGAUUAUGAUUGGGACACAUGGU-GAAAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAUGGCCAGCAGGUCCU ..........--.............(((((.(((...-...)))---.(((-----------------(((((((.(((....((((.....)))).--..))).))))))))))))))) ( -35.50) >DroYak_CAF1 10206 95 + 1 GCGAACCUUU--UUUGAUUAUGAUUGGGACACAUGGC-GAAAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAAAGCCAGCAGGUCCU ..........--.............(((((.(((...-...)))---.(((-----------------(((((...(((....((((.....)))).--..)))...))))))))))))) ( -30.30) >DroPer_CAF1 41830 120 + 1 GCGGACCUUUUUUUUGAUUAUGAUUGGAACACAUGGGCGACACAGGGUUUGGGGGGUAGGGGAGGGGAGGGGCUGACUCAAUAAGGCUAAAAGCCAGCGACGAGGACGCCACGAGGUCCU ..(((((((((((((..((((..((.((((...(((....).))..)))).))..))))..)))))))..(((...(((.....(((.....)))......)))...)))...)))))). ( -33.00) >consensus GCGAACCUUU__UUUGAUUAUGAUUGGGACACAUGGU_GAAAUG___ACUG_________________CUGGUCAACUCAAUAUGGCUAAAAGCCAA__GCGAGAAGGCCAGCAGGUCCU ..........................((((....((((........))))..................(((((...(((....((((.....)))).....)))...)))))...)))). (-16.34 = -16.45 + 0.11)

| Location | 12,963,913 – 12,964,008 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12963913 95 - 22407834 AGGACCUGCUGGCCAUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUUUC-ACCAUGUGUCCCAAUCAUAAUCAAA--AAAGGUUCGC .(((((((((((.((.((((.--.((((.....))))...)))).)).))))-----------------)...---......-....((((.......))))......--..)))))).. ( -30.00) >DroPse_CAF1 43482 120 - 1 AGGACCUCGUGGCGUCCUCGUCGCUGGCUUUUAGCCUUAUUGAGUCAGCCCCUCCCCUCCCCUACCCCCCAAACCCUGUGUCGCCCAUGUGUUCGAAUCAUAAUCAAAAAAAAGGUCCGC .((((((...((((....))))((((((((...........))))))))...........................(((((((..(....)..)))..))))..........)))))).. ( -27.00) >DroSec_CAF1 8706 95 - 1 AGGACCUGCUGGCCAACUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUUUC-ACCAUGUGUCCCAAUCAUAAUCAAA--AAAGGUUCGC .(((((((((((.(((((((.--.((((.....))))...))))))).))))-----------------)...---......-....((((.......))))......--..)))))).. ( -34.10) >DroSim_CAF1 9522 95 - 1 AGGACCUGCUGGCCAUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUUUC-ACCAUGUGUCCCAAUCAUAAUCAAA--AAAGGUUCGC .(((((((((((.((.((((.--.((((.....))))...)))).)).))))-----------------)...---......-....((((.......))))......--..)))))).. ( -30.00) >DroYak_CAF1 10206 95 - 1 AGGACCUGCUGGCUUUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUUUC-GCCAUGUGUCCCAAUCAUAAUCAAA--AAAGGUUCGC .((((((((((((...((((.--.((((.....))))...))))..).))))-----------------)...---......-....((((.......))))......--..)))))).. ( -28.00) >DroPer_CAF1 41830 120 - 1 AGGACCUCGUGGCGUCCUCGUCGCUGGCUUUUAGCCUUAUUGAGUCAGCCCCUCCCCUCCCCUACCCCCCAAACCCUGUGUCGCCCAUGUGUUCCAAUCAUAAUCAAAAAAAAGGUCCGC .((((((.(.((((.(...(..((((((((...........))))))))..).................((.....)).).))))).((((.......))))..........)))))).. ( -27.00) >consensus AGGACCUGCUGGCCAUCUCGC__UUGGCUUUUAGCCAUAUUGAGUUGACCAG_________________CAGU___CAUUUC_ACCAUGUGUCCCAAUCAUAAUCAAA__AAAGGUUCGC .((((((.((((....((((....((((.....))))...))))....))))...................................((((.......))))..........)))))).. (-21.08 = -21.08 + 0.00)

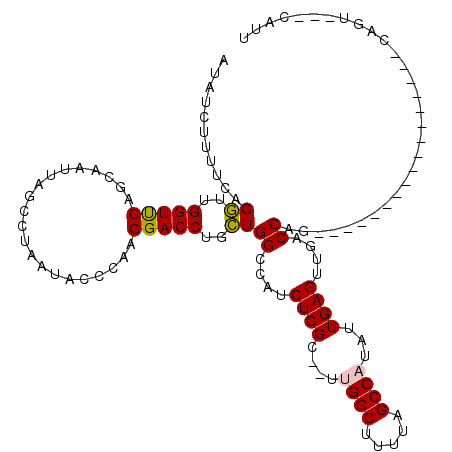
| Location | 12,963,950 – 12,964,048 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12963950 98 + 22407834 AAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAUGGCCAGCAGGUCCUUGGGUAUUAGGCUAAUUGCUGAACCAACCUGAAAAGAUAU ....---.(((-----------------(((((((.(((....((((.....)))).--..))).))))))))))(((.(..(((....(((.....)))......)))..)...))).. ( -36.00) >DroPse_CAF1 43522 120 + 1 CACAGGGUUUGGGGGGUAGGGGAGGGGAGGGGCUGACUCAAUAAGGCUAAAAGCCAGCGACGAGGACGCCACGAGGUCCUUGGGUAUUAGGCUAAUUGUCGGACCAAGCUGAAAAGAUAU .....(((((((......(((..((.......))..))).....(((....((((.((..(((((((........))))))).))....))))....)))...))))))).......... ( -32.10) >DroSec_CAF1 8743 98 + 1 AAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGUUGGCCAGCAGGUCCUUGGGUAUUAGGCUAAUUGCUGAACCAACCUGAAAAGAUAU ....---.(((-----------------(((((((((((....((((.....)))).--..))))))))))))))(((.(..(((....(((.....)))......)))..)...))).. ( -40.20) >DroSim_CAF1 9559 98 + 1 AAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAUGGCCAGCAGGUCCUUGGGUAUUAGGCUAAUUGCUGAACCAACCUGAAAAGAUAU ....---.(((-----------------(((((((.(((....((((.....)))).--..))).))))))))))(((.(..(((....(((.....)))......)))..)...))).. ( -36.00) >DroYak_CAF1 10243 98 + 1 AAUG---ACUG-----------------CUGGUCAACUCAAUAUGGCUAAAAGCCAA--GCGAGAAAGCCAGCAGGUCCUUGGGUAUUAGGCUAAUUCCUGAACCAACCUGAAAAGAUAU ....---.(((-----------------(((((...(((....((((.....)))).--..)))...))))))))(((.(..(((.(((((......)))))....)))..)...))).. ( -30.80) >DroPer_CAF1 41870 120 + 1 CACAGGGUUUGGGGGGUAGGGGAGGGGAGGGGCUGACUCAAUAAGGCUAAAAGCCAGCGACGAGGACGCCACGAGGUCCUUGGGUAUUAGGCUAAUUGUCGGACCAAGCUGAAAAGAUAU .....(((((((......(((..((.......))..))).....(((....((((.((..(((((((........))))))).))....))))....)))...))))))).......... ( -32.10) >consensus AAUG___ACUG_________________CUGGUCAACUCAAUAUGGCUAAAAGCCAA__GCGAGAAGGCCAGCAGGUCCUUGGGUAUUAGGCUAAUUGCUGAACCAACCUGAAAAGAUAU ............................(((((...(((....((((.....)))).....)))...)))))(((((.....(((....(((.....)))..))).)))))......... (-20.27 = -20.67 + 0.39)



| Location | 12,963,950 – 12,964,048 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12963950 98 - 22407834 AUAUCUUUUCAGGUUGGUUCAGCAAUUAGCCUAAUACCCAAGGACCUGCUGGCCAUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUU ..........(((((((((....)))))))))..........(((.((((((.((.((((.--.((((.....))))...)))).)).))))-----------------))))---)... ( -33.40) >DroPse_CAF1 43522 120 - 1 AUAUCUUUUCAGCUUGGUCCGACAAUUAGCCUAAUACCCAAGGACCUCGUGGCGUCCUCGUCGCUGGCUUUUAGCCUUAUUGAGUCAGCCCCUCCCCUCCCCUACCCCCCAAACCCUGUG ...........((..(((((.....................)))))..))((((....))))((((((((...........))))))))............................... ( -22.90) >DroSec_CAF1 8743 98 - 1 AUAUCUUUUCAGGUUGGUUCAGCAAUUAGCCUAAUACCCAAGGACCUGCUGGCCAACUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUU ..........(((((((((....)))))))))..........(((.((((((.(((((((.--.((((.....))))...))))))).))))-----------------))))---)... ( -37.50) >DroSim_CAF1 9559 98 - 1 AUAUCUUUUCAGGUUGGUUCAGCAAUUAGCCUAAUACCCAAGGACCUGCUGGCCAUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUU ..........(((((((((....)))))))))..........(((.((((((.((.((((.--.((((.....))))...)))).)).))))-----------------))))---)... ( -33.40) >DroYak_CAF1 10243 98 - 1 AUAUCUUUUCAGGUUGGUUCAGGAAUUAGCCUAAUACCCAAGGACCUGCUGGCUUUCUCGC--UUGGCUUUUAGCCAUAUUGAGUUGACCAG-----------------CAGU---CAUU ..........(((((((((....)))))))))..........(((.(((((((...((((.--.((((.....))))...))))..).))))-----------------))))---)... ( -32.50) >DroPer_CAF1 41870 120 - 1 AUAUCUUUUCAGCUUGGUCCGACAAUUAGCCUAAUACCCAAGGACCUCGUGGCGUCCUCGUCGCUGGCUUUUAGCCUUAUUGAGUCAGCCCCUCCCCUCCCCUACCCCCCAAACCCUGUG ...........((..(((((.....................)))))..))((((....))))((((((((...........))))))))............................... ( -22.90) >consensus AUAUCUUUUCAGGUUGGUUCAGCAAUUAGCCUAAUACCCAAGGACCUGCUGGCCAUCUCGC__UUGGCUUUUAGCCAUAUUGAGUUGACCAG_________________CAGU___CAUU ...........((..(((((.....................)))))..))((....((((....((((.....))))...))))....)).............................. (-16.65 = -16.32 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:53 2006