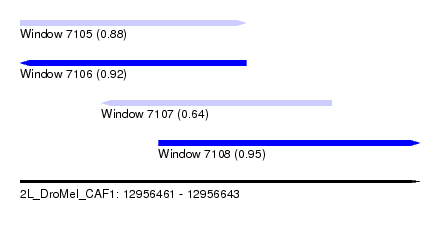
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,956,461 – 12,956,643 |
| Length | 182 |
| Max. P | 0.952008 |

| Location | 12,956,461 – 12,956,564 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12956461 103 + 22407834 UUUUAUUUUUGCAGCCAAUUAAUAUUGAAAUAAAUUCAAAUGACAUGGUUAAAAGCCAACC--------------AGCAUGUGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGC .......(((((((((((((((...((((((.....((.(((...(((((.......))))--------------).))).))))))))..)))).))))))))))).......... ( -27.30) >DroSec_CAF1 1206 117 + 1 UUUAUUUUUUUCAGCCAAUUAAUAUUGAAAUAAAUUCAAAUGACACGGUUAAAAGCCAACCAGCUUCAUAUGGCCAGCAUGUGGUUUUACUUUAAUUUGGCUGCAAACUUUGCACGC .......(((.(((((((.....((((((.((((..((.(((....(((((.((((......))))....)))))..))).))..)))).))))))))))))).))).......... ( -22.60) >DroSim_CAF1 2005 116 + 1 UUUAUUU-UUGCAGCCAAUUAAUAUUGAAAUAAAUUCAAAUGACAUGGUUAAAAGCCAACCAGAUUCAUAUGGCCAGCAUGCGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGC ......(-((((((((((((((...((((((.....((.((((..(((((.......)))))...)))).))((......)).))))))..)))).))))))))))).......... ( -30.70) >consensus UUUAUUUUUUGCAGCCAAUUAAUAUUGAAAUAAAUUCAAAUGACAUGGUUAAAAGCCAACCAG_UUCAUAUGGCCAGCAUGUGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGC ........((((((((((((((...((((((....((....))...(((.....)))..........................))))))..)))).))))))))))........... (-20.55 = -20.67 + 0.11)

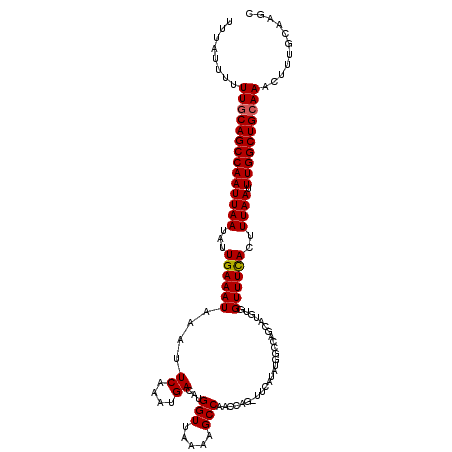
| Location | 12,956,461 – 12,956,564 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -22.80 |
| Energy contribution | -25.00 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12956461 103 - 22407834 GCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCACAUGCU--------------GGUUGGCUUUUAACCAUGUCAUUUGAAUUUAUUUCAAUAUUAAUUGGCUGCAAAAAUAAAA ..........(((((((((((.((((.(((....)))(((((--------------(((((.....)))))).).)))(((((.....)))))..)))))))))))))))....... ( -26.80) >DroSec_CAF1 1206 117 - 1 GCGUGCAAAGUUUGCAGCCAAAUUAAAGUAAAACCACAUGCUGGCCAUAUGAAGCUGGUUGGCUUUUAACCGUGUCAUUUGAAUUUAUUUCAAUAUUAAUUGGCUGAAAAAAAUAAA .........((((.(((((((.((((.............((..((((........))))..))...............(((((.....)))))..)))))))))))....))))... ( -25.00) >DroSim_CAF1 2005 116 - 1 GCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCGCAUGCUGGCCAUAUGAAUCUGGUUGGCUUUUAACCAUGUCAUUUGAAUUUAUUUCAAUAUUAAUUGGCUGCAA-AAAUAAA ..........(((((((((((.((((((((....)))..((..((((........))))..)).))))).........(((((.....)))))......))))))))))-)...... ( -32.40) >consensus GCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCACAUGCUGGCCAUAUGAA_CUGGUUGGCUUUUAACCAUGUCAUUUGAAUUUAUUUCAAUAUUAAUUGGCUGCAAAAAAUAAA ..........(((((((((((.((((((((....)))..((((((((........)))))))).))))).........(((((.....)))))......)))))))))))....... (-22.80 = -25.00 + 2.20)

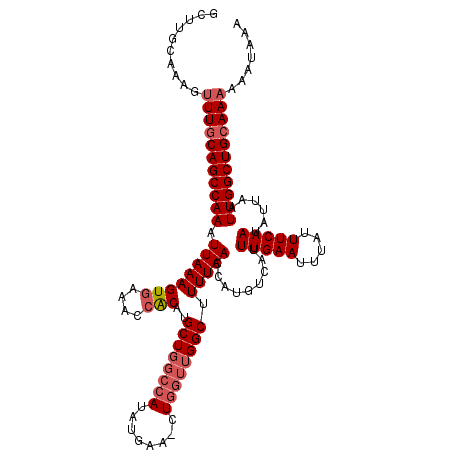
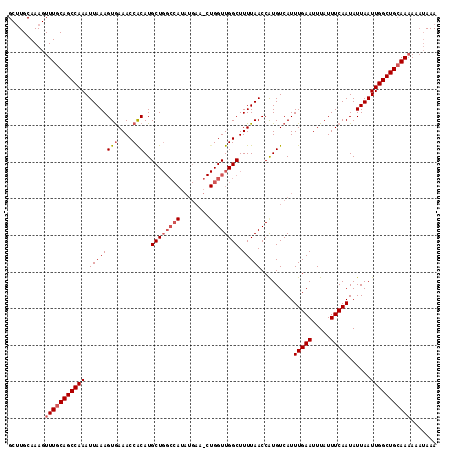
| Location | 12,956,498 – 12,956,603 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.91 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.95 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12956498 105 - 22407834 UUUCGAACAUCCUUGGCUGAAACUGCCCCCA-AACAGUGGGCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCACAUGCU--------------GGUUGGCUUUUAACCAUGUCAUUU ....((.(((..((((((((((((((.((((-.....))))...))..))))).)))))))...(((((..(((((.....)--------------)))).))))).....))))).... ( -28.10) >DroSec_CAF1 1243 119 - 1 UUUCGAACAUCCUUGGCUGAAACUGCCCCCA-AACAGUGGGCGUGCAAAGUUUGCAGCCAAAUUAAAGUAAAACCACAUGCUGGCCAUAUGAAGCUGGUUGGCUUUUAACCGUGUCAUUU ....((.(((..(((((((((((((((((((-.....)))).).))..))))).)))))))..................((..((((........))))..))........))))).... ( -33.80) >DroSim_CAF1 2041 119 - 1 UUUCGAACAUCCUUGGCUGAAACUGCCCCCA-AACAGUGGGCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCGCAUGCUGGCCAUAUGAAUCUGGUUGGCUUUUAACCAUGUCAUUU ....((.(((..((((((((((((((.((((-.....))))...))..))))).))))))).((((((((....)))..((..((((........))))..)).)))))..))))).... ( -33.70) >DroYak_CAF1 1400 113 - 1 UUUCGAACAUCCUUGGCUGGAACUGCCCCCAAAACAGUGGGCUUGCAAAGUGUGCAGCCAAAUUAAAGUGAAACCACAUGCUGGCAAUAUUAGGCUGGUUGG-------UCAUGUCACCU ............(((((((.((((((.((((......))))...))..))).).)))))))......((((.((((...((..((........))..)))))-------)....)))).. ( -31.10) >consensus UUUCGAACAUCCUUGGCUGAAACUGCCCCCA_AACAGUGGGCUUGCAAAGUUUGCAGCCAAAUUAAAGUGAAACCACAUGCUGGCCAUAUGAAGCUGGUUGGCUUUUAACCAUGUCAUUU ....((.(((..((((((((((((((.((((......))))...))..))))).)))))))............(((...((((((........))))))))).........))))).... (-24.83 = -24.95 + 0.12)

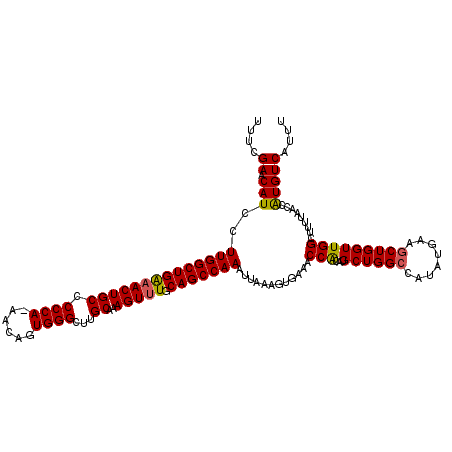
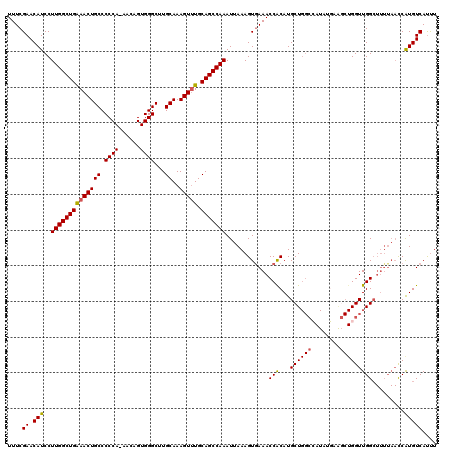
| Location | 12,956,524 – 12,956,643 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -36.29 |
| Energy contribution | -37.10 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12956524 119 + 22407834 CAUGUGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGCCCACUGUU-UGGGGGCAGUUUCAGCCAAGGAUGUUCGAAAUGAAAUUCGGCCGUCAAGGUGAUUGUUUUCAAACCUCAAG ..((.(((((((((((..((((((((.(((((..((...((((.....-)))).)))))))))))))))((((..((((......))))..))))))))))).........)))).)).. ( -40.40) >DroSec_CAF1 1283 119 + 1 CAUGUGGUUUUACUUUAAUUUGGCUGCAAACUUUGCACGCCCACUGUU-UGGGGGCAGUUUCAGCCAAGGAUGUUCGAAAUGAAACUCGGCCGUCAAGGUGAUUGUUUUCAAAACUCAAG ..((.(((((((((((..((((((((.(((((..((.(.((((.....-))))))))))))))))))))((((..(((........)))..)))))))))((......)))))))))).. ( -35.50) >DroSim_CAF1 2081 119 + 1 CAUGCGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGCCCACUGUU-UGGGGGCAGUUUCAGCCAAGGAUGUUCGAAAUGAAAUUCGGCCGUCAAGGUGAUUGUUUUCAAACCUCAAG ..((.(((((((((((..((((((((.(((((..((...((((.....-)))).)))))))))))))))((((..((((......))))..))))))))))).........)))).)).. ( -40.10) >DroYak_CAF1 1433 120 + 1 CAUGUGGUUUCACUUUAAUUUGGCUGCACACUUUGCAAGCCCACUGUUUUGGGGGCAGUUCCAGCCAAGGAUGUUCGAAAUGAAAUUCGGCCGUCAAGGUGAUUGUUUUCAAACCUCAAG ..((.(((((((((((..((((((((((.....)))..((((.((.....))))))......)))))))((((..((((......))))..))))))))))).........)))).)).. ( -38.10) >consensus CAUGUGGUUUCACUUUAAUUUGGCUGCAAACUUUGCAAGCCCACUGUU_UGGGGGCAGUUUCAGCCAAGGAUGUUCGAAAUGAAAUUCGGCCGUCAAGGUGAUUGUUUUCAAACCUCAAG ..((.(((((((((((..((((((((.(((((..((...((((......)))).)))))))))))))))((((..((((......))))..))))))))))).........)))).)).. (-36.29 = -37.10 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:47 2006