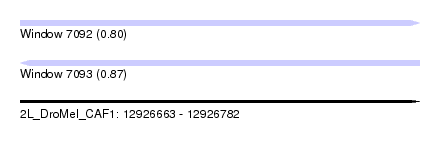
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,926,663 – 12,926,782 |
| Length | 119 |
| Max. P | 0.870246 |

| Location | 12,926,663 – 12,926,782 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12926663 119 + 22407834 UCUCCCUGCCCCUGCAGAAAGACAUUCAGGGCAGGAUCGUAAUGGCAAAGCAAGG-UAUUCACUCCUGGACCGCCAUGGAGCGCACAAUGGCCAUACAGAUCCAUCCCGAUGUCUCCAUC .....((((....))))..(((((((..(((..(((((...(((((....((..(-(...(.(((((((....))).)))).).))..)))))))...)))))..))))))))))..... ( -38.20) >DroSec_CAF1 105841 119 + 1 UCUCGCUGCCCCUGCAGAAACACAUUCAGGGCAGGAUCAUAAUGGCCAAGCAAGG-UAUUCACUCCUGGACCGCCAUGGAGCGCACAAUGGCCAUACAGAUCCAUCCCGAUGUCUCCAUC .....((((....))))....(((((..(((..(((((...(((((((.((...(-....).(((((((....))).)))).))....)))))))...)))))..))))))))....... ( -36.50) >DroSim_CAF1 109850 119 + 1 UCUCGCUGCCCCUGCAGAAAGACAUUCAGGGCAGGAUCAUAAUGGCCAAGCAAGG-UAUUCACUCCUGGACCGCCAUGGAGCGCAGAAUGGCCAUACAGAUCCAUCCCGAUGUCUCCAUC .....((((....))))..(((((((..(((..(((((...(((((((..(...(-(.....(((((((....))).)))).)).)..)))))))...)))))..))))))))))..... ( -41.20) >DroEre_CAF1 113012 119 + 1 UCUCGCUGCCCCUGCAGAAGCACAUUCAGGGCAGGAUUAUAAUGGCCAAUCAAGG-UGUUCACUCCUGGACCGCCUUGGAGCGCACAAUGGCCAUCCAAAUCCAUCCCGAUGUCUCCAUC .....((((....)))).((.(((((..(((..(((((...(((((((.((((((-((((((....)))).)))))))).(....)..)))))))...)))))..))))))))))..... ( -38.20) >DroYak_CAF1 108210 119 + 1 UUUCGCUGCCCCUGCAGAAAAACAUUCAGGGCAAGAUCGUAAUGGCCAGGCAAGG-CAUUCACUCCUGGACAGCCAUGGAGCGCACAAUGGCCAUACAGAUCCAUCCCGAUGUCUCCAUC (((..((((....))))..)))......(((...((((...(((((((.((..(.-....).(((((((....))).)))).))....)))))))...))))...)))((((....)))) ( -36.30) >DroAna_CAF1 126287 113 + 1 UAUCAGAGCAGUUGCAAAGGGUAAUCCAAAACAGGAUUA-----GCCAGGCCGGGGCGCGGAGUUU--UAGCGUUUUGGACCGCAGCAUGACCAUCCAGGUGCACCCGGAUGUCAGCAUC .......((.(((((...((.((((((......))))))-----.)).(((((((((((.......--..))))))))).))))))).((((.((((.((....)).))))))))))... ( -48.50) >consensus UCUCGCUGCCCCUGCAGAAAGACAUUCAGGGCAGGAUCAUAAUGGCCAAGCAAGG_UAUUCACUCCUGGACCGCCAUGGAGCGCACAAUGGCCAUACAGAUCCAUCCCGAUGUCUCCAUC .....((((....))))...........(((..(((((...(((((((.((((.....))..(((((((....))).)))).))....)))))))...)))))..)))((((....)))) (-23.67 = -24.90 + 1.23)

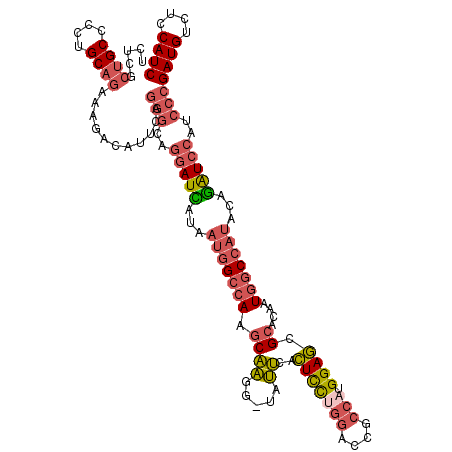
| Location | 12,926,663 – 12,926,782 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -46.25 |
| Consensus MFE | -32.58 |
| Energy contribution | -34.53 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12926663 119 - 22407834 GAUGGAGACAUCGGGAUGGAUCUGUAUGGCCAUUGUGCGCUCCAUGGCGGUCCAGGAGUGAAUA-CCUUGCUUUGCCAUUACGAUCCUGCCCUGAAUGUCUUUCUGCAGGGGCAGGGAGA ((((....))))(((..(((((.(((((((((..(((((((((.(((....)))))))))..))-)..))....)))).))))))))..)))......(((..((((....))))..))) ( -50.00) >DroSec_CAF1 105841 119 - 1 GAUGGAGACAUCGGGAUGGAUCUGUAUGGCCAUUGUGCGCUCCAUGGCGGUCCAGGAGUGAAUA-CCUUGCUUGGCCAUUAUGAUCCUGCCCUGAAUGUGUUUCUGCAGGGGCAGCGAGA ((((....)))).....(((((...(((((((..(((((((((.(((....)))))))))..))-)......)))))))...)))))((((((...(((......)))))))))...... ( -47.70) >DroSim_CAF1 109850 119 - 1 GAUGGAGACAUCGGGAUGGAUCUGUAUGGCCAUUCUGCGCUCCAUGGCGGUCCAGGAGUGAAUA-CCUUGCUUGGCCAUUAUGAUCCUGCCCUGAAUGUCUUUCUGCAGGGGCAGCGAGA ((((....))))(((..(((((...(((((((..(..((((((.(((....)))))))))....-....)..)))))))...)))))..)))......((((.((((....)))).)))) ( -50.60) >DroEre_CAF1 113012 119 - 1 GAUGGAGACAUCGGGAUGGAUUUGGAUGGCCAUUGUGCGCUCCAAGGCGGUCCAGGAGUGAACA-CCUUGAUUGGCCAUUAUAAUCCUGCCCUGAAUGUGCUUCUGCAGGGGCAGCGAGA ((((....))))(((..(((((..((((((((.....((((....))))(((.(((.(....).-))).)))))))))))..)))))..)))......((((((....))))))...... ( -46.80) >DroYak_CAF1 108210 119 - 1 GAUGGAGACAUCGGGAUGGAUCUGUAUGGCCAUUGUGCGCUCCAUGGCUGUCCAGGAGUGAAUG-CCUUGCCUGGCCAUUACGAUCUUGCCCUGAAUGUUUUUCUGCAGGGGCAGCGAAA ((((....))))(((..(((((.(((((((((.....((((((.(((....)))))))))...(-(...)).)))))).))))))))..)))......((((.((((....)))).)))) ( -45.90) >DroAna_CAF1 126287 113 - 1 GAUGCUGACAUCCGGGUGCACCUGGAUGGUCAUGCUGCGGUCCAAAACGCUA--AAACUCCGCGCCCCGGCCUGGC-----UAAUCCUGUUUUGGAUUACCCUUUGCAACUGCUCUGAUA ...(((((((((((((....))))))).)))).))(((((((.....(((..--.......)))....)))).((.-----((((((......)))))).))...)))............ ( -36.50) >consensus GAUGGAGACAUCGGGAUGGAUCUGUAUGGCCAUUGUGCGCUCCAUGGCGGUCCAGGAGUGAAUA_CCUUGCUUGGCCAUUAUGAUCCUGCCCUGAAUGUCUUUCUGCAGGGGCAGCGAGA ((((....))))(((..(((((...(((((((.(((.((((((.(((....))))))))).)))........)))))))...)))))..)))...........((((....))))..... (-32.58 = -34.53 + 1.95)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:32 2006