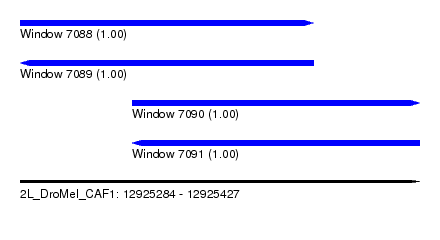
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,925,284 – 12,925,427 |
| Length | 143 |
| Max. P | 0.998807 |

| Location | 12,925,284 – 12,925,389 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12925284 105 + 22407834 AAUUCUUCUCAAUAUGGAAUUCGAAUCGCAAAUGCAUAUUGAAGGCUUCGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGUCUGUAAAAU-----------UUGAGAUGCGUAGCCU ........((((((((.(.((((...)).)).).))))))))(((((.(((.(((((((((((((((.((...----....))))).))))))-----------)))))).))).))))) ( -32.30) >DroSec_CAF1 104461 105 + 1 AAAUCUUCAUAAUAUGGAGUUCGAAUGGCAAAUGCAUAUUGAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGACUGUAAAAU-----------UUGAGAUGCGUAGCCU ....((((((...))))))(((((...((....))...)))))((((((((.((((((((((((((.(((...----....))))).))))))-----------)))))).)))))))). ( -36.60) >DroSim_CAF1 108471 105 + 1 AAAUCUUCAUAAUAUGGAAUUAGAAUGGCAAAUGCAUAUUGAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGACUGUAAAAU-----------UUGAGAUGCGUAGCCU .........(((((((..(((.(.....).))).))))))).(((((((((.((((((((((((((.(((...----....))))).))))))-----------)))))).))))))))) ( -33.10) >DroEre_CAF1 111231 105 + 1 AAUUAGUCACAAUAUGGAAAUUGAAUCGCAACUGCAUAUUGUUGGCUACGUCUCUCAGAUUUUGAGUUCGUAG----UUCUUGACUGAAAAAU-----------UUGAGAUGCGUAGCCA ........((((((((.(..(((.....))).).))))))))(((((((((.(((((((((((.(((.((...----....)))))..)))))-----------)))))).))))))))) ( -35.80) >DroYak_CAF1 106513 118 + 1 --UUUAUCACAAUAUGGAAAUCGAAUCGCAAGUGCAUAUUGUUGGCUACGUCUCUCAGAUUUUGAGUUCGUAGUCAGUCCGUGGCUGAAAAAUUUGAGCAAAAUUUGAGAUGCGUAGCCA --......((((((((.(...((...))....).))))))))(((((((((.((((((((((((...(((...(((((.....)))))......))).)))))))))))).))))))))) ( -43.40) >consensus AAUUCUUCACAAUAUGGAAUUCGAAUCGCAAAUGCAUAUUGAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG____UUCUUGACUGUAAAAU___________UUGAGAUGCGUAGCCU ........((((((((.(..............).))))))))(((((((((.((((((((((((...(((...........)))...))))))...........)))))).))))))))) (-23.52 = -23.48 + -0.04)


| Location | 12,925,284 – 12,925,389 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12925284 105 - 22407834 AGGCUACGCAUCUCAA-----------AUUUUACAGACAAGAA----CUACGAUCUCAAAAUUUGAGAAACGAAGCCUUCAAUAUGCAUUUGCGAUUCGAAUUCCAUAUUGAGAAGAAUU .((((.((..((((((-----------(((((..(((...(..----...)..))).)))))))))))..)).))))(((((((((.(((((.....)))))..)))))))))....... ( -27.50) >DroSec_CAF1 104461 105 - 1 AGGCUACGCAUCUCAA-----------AUUUUACAGUCAAGAA----CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUCAAUAUGCAUUUGCCAUUCGAACUCCAUAUUAUGAAGAUUU .(((((((..((((((-----------(((((...(((.....----....)))...)))))))))))..)))))))(((((((((..((((.....))))...)))))..))))..... ( -27.40) >DroSim_CAF1 108471 105 - 1 AGGCUACGCAUCUCAA-----------AUUUUACAGUCAAGAA----CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUCAAUAUGCAUUUGCCAUUCUAAUUCCAUAUUAUGAAGAUUU .(((((((..((((((-----------(((((...(((.....----....)))...)))))))))))..)))))))(((((((((...(((......)))...)))))..))))..... ( -26.60) >DroEre_CAF1 111231 105 - 1 UGGCUACGCAUCUCAA-----------AUUUUUCAGUCAAGAA----CUACGAACUCAAAAUCUGAGAGACGUAGCCAACAAUAUGCAGUUGCGAUUCAAUUUCCAUAUUGUGACUAAUU ((((((((..(((((.-----------(((((....((.....----....))....))))).)))))..))))))))((((((((.(((((.....)))))..))))))))........ ( -31.00) >DroYak_CAF1 106513 118 - 1 UGGCUACGCAUCUCAAAUUUUGCUCAAAUUUUUCAGCCACGGACUGACUACGAACUCAAAAUCUGAGAGACGUAGCCAACAAUAUGCACUUGCGAUUCGAUUUCCAUAUUGUGAUAAA-- ((((((((..(((((.((((((..........(((((....).)))).........)))))).)))))..))))))))((((((((......((...)).....))))))))......-- ( -34.81) >consensus AGGCUACGCAUCUCAA___________AUUUUACAGUCAAGAA____CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUCAAUAUGCAUUUGCGAUUCGAAUUCCAUAUUGUGAAGAAUU ((((((((..(((((................................................)))))..))))))))((((((((.(((((.....)))))..))))))))........ (-19.65 = -20.25 + 0.60)

| Location | 12,925,324 – 12,925,427 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12925324 103 + 22407834 GAAGGCUUCGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGUCUGUAAAAU-----------UUGAGAUGCGUAGCCUUACAAAAUGAUGUGG--ACUCAAAUCUUGUAAAAAUUAUG .((((((.(((.(((((((((((((((.((...----....))))).))))))-----------)))))).))).))))))((((...(((.((.--...)).))))))).......... ( -28.50) >DroSec_CAF1 104501 103 + 1 GAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGACUGUAAAAU-----------UUGAGAUGCGUAGCCUUUCAAAAUGAUGUGG--ACUUAAAUCUUGUAAAAAUUAUU (((((((((((.((((((((((((((.(((...----....))))).))))))-----------)))))).)))))))))))...((((((....--((.........))....)))))) ( -33.50) >DroSim_CAF1 108511 103 + 1 GAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG----UUCUUGACUGUAAAAU-----------UUGAGAUGCGUAGCCUUACAAAAUGAUGUGG--ACUCAAAUCUUGUAAAAAUUAUG .((((((((((.((((((((((((((.(((...----....))))).))))))-----------)))))).))))))))))((((...(((.((.--...)).))))))).......... ( -33.10) >DroEre_CAF1 111271 105 + 1 GUUGGCUACGUCUCUCAGAUUUUGAGUUCGUAG----UUCUUGACUGAAAAAU-----------UUGAGAUGCGUAGCCAUACAAAAUGAUGUAAAAACUGGAACCUUGUAAAAAUUAGU ..(((((((((.(((((((((((.(((.((...----....)))))..)))))-----------)))))).)))))))))((((......)))).......................... ( -30.90) >DroYak_CAF1 106551 112 + 1 GUUGGCUACGUCUCUCAGAUUUUGAGUUCGUAGUCAGUCCGUGGCUGAAAAAUUUGAGCAAAAUUUGAGAUGCGUAGCCAUACAAAAUGA--------CUUGAAUCUUGUACAAAUUAUU ..(((((((((.((((((((((((...(((...(((((.....)))))......))).)))))))))))).)))))))))(((((.((..--------.....)).)))))......... ( -39.60) >consensus GAAGGCUACGUUUCUCAAAUUUUGAGAUCGUAG____UUCUUGACUGUAAAAU___________UUGAGAUGCGUAGCCUUACAAAAUGAUGUGG__ACUCAAAUCUUGUAAAAAUUAUU .((((((((((.((((((((((((...(((...........)))...))))))...........)))))).))))))))))....................................... (-19.94 = -20.30 + 0.36)


| Location | 12,925,324 – 12,925,427 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.37 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12925324 103 - 22407834 CAUAAUUUUUACAAGAUUUGAGU--CCACAUCAUUUUGUAAGGCUACGCAUCUCAA-----------AUUUUACAGACAAGAA----CUACGAUCUCAAAAUUUGAGAAACGAAGCCUUC ........((((((((..(((..--.....)))))))))))((((.((..((((((-----------(((((..(((...(..----...)..))).)))))))))))..)).))))... ( -24.30) >DroSec_CAF1 104501 103 - 1 AAUAAUUUUUACAAGAUUUAAGU--CCACAUCAUUUUGAAAGGCUACGCAUCUCAA-----------AUUUUACAGUCAAGAA----CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUC ...........((((((......--.......)))))).(((((((((..((((((-----------(((((...(((.....----....)))...)))))))))))..))))))))). ( -26.72) >DroSim_CAF1 108511 103 - 1 CAUAAUUUUUACAAGAUUUGAGU--CCACAUCAUUUUGUAAGGCUACGCAUCUCAA-----------AUUUUACAGUCAAGAA----CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUC ........((((((((..(((..--.....)))))))))))(((((((..((((((-----------(((((...(((.....----....)))...)))))))))))..)))))))... ( -30.00) >DroEre_CAF1 111271 105 - 1 ACUAAUUUUUACAAGGUUCCAGUUUUUACAUCAUUUUGUAUGGCUACGCAUCUCAA-----------AUUUUUCAGUCAAGAA----CUACGAACUCAAAAUCUGAGAGACGUAGCCAAC .........((((((((....((....))...))))))))((((((((..(((((.-----------(((((....((.....----....))....))))).)))))..)))))))).. ( -26.10) >DroYak_CAF1 106551 112 - 1 AAUAAUUUGUACAAGAUUCAAG--------UCAUUUUGUAUGGCUACGCAUCUCAAAUUUUGCUCAAAUUUUUCAGCCACGGACUGACUACGAACUCAAAAUCUGAGAGACGUAGCCAAC ........(((((((((.....--------..)))))))))(((((((..(((((.((((((..........(((((....).)))).........)))))).)))))..)))))))... ( -33.61) >consensus AAUAAUUUUUACAAGAUUUAAGU__CCACAUCAUUUUGUAAGGCUACGCAUCUCAA___________AUUUUACAGUCAAGAA____CUACGAUCUCAAAAUUUGAGAAACGUAGCCUUC .........((((((((...............))))))))((((((((..(((((................................................)))))..)))))))).. (-18.61 = -18.37 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:30 2006