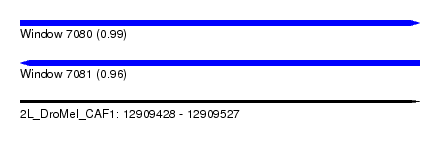
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,909,428 – 12,909,527 |
| Length | 99 |
| Max. P | 0.994808 |

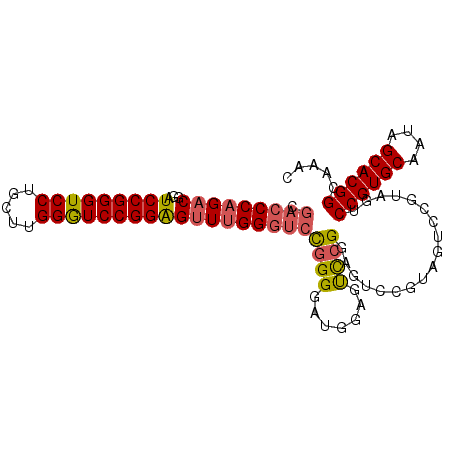
| Location | 12,909,428 – 12,909,527 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -47.55 |
| Consensus MFE | -34.99 |
| Energy contribution | -35.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
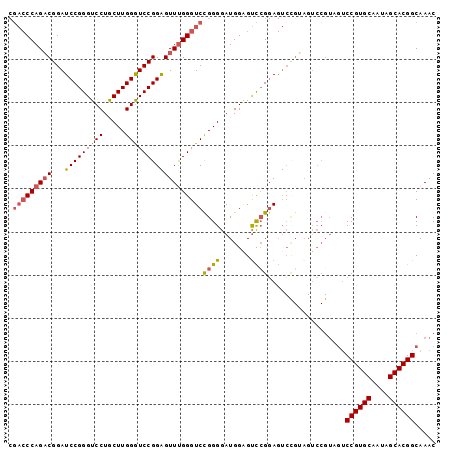
>2L_DroMel_CAF1 12909428 99 + 22407834 CGACCCAGACGGAUCCGGGUCCUGCUUGGGUCCGGAGUUUGGGUCCGGGGAUGGAGUCCGGAGUCCGUAGUCCGUAGUCCGUGCAAUAGCACGGCAAAC .(((((((((...(((((..((.....))..))))))))))))))((((.(((((.(....).)))))..))))..(.((((((....))))))).... ( -51.10) >DroSec_CAF1 87081 85 + 1 CGACCCAGACGGAUCCGGGUCCUACUUGGGUCCGGAGUUUGGGUCCGGGGAUGGUGUCCGGAGU--------------CCGUGCAAUAGCACGGCAAAC .(((((((((...(((((..((.....))..))))))))))))))((((.......))))..(.--------------((((((....))))))).... ( -44.00) >DroSim_CAF1 92155 99 + 1 CGACCCAGACGGAUCCGGGUCCUGCUUGGGUCCGGGGUUUGGGUCUGGGGAUGGGGUCCGGAGUCCGUAGUCCGUAGUCCGUGCAAUAGCACGGCAAAC .(((((((((...(((((..((.....))..)))))))))))))).(((.(((((.(....).)))))..)))...(.((((((....))))))).... ( -48.10) >DroEre_CAF1 92246 92 + 1 ----CCAGCCGGAUCCGGGUCCUGCUUGGAUCCGGAGUUCGGGUCCGGGGAUGGAGCUGG-A--CUGGAGUCCGUAGUCCGUGCAAUAGCACGGCAAAC ----((..(((((((((((.....)))))))))))(((((((.((((....)))).))))-)--))))........(.((((((....))))))).... ( -47.00) >consensus CGACCCAGACGGAUCCGGGUCCUGCUUGGGUCCGGAGUUUGGGUCCGGGGAUGGAGUCCGGAGUCCGUAGUCCGUAGUCCGUGCAAUAGCACGGCAAAC .(((((((((...(((((((((.....))))))))))))))))))((((.......))))..................((((((....))))))..... (-34.99 = -35.55 + 0.56)

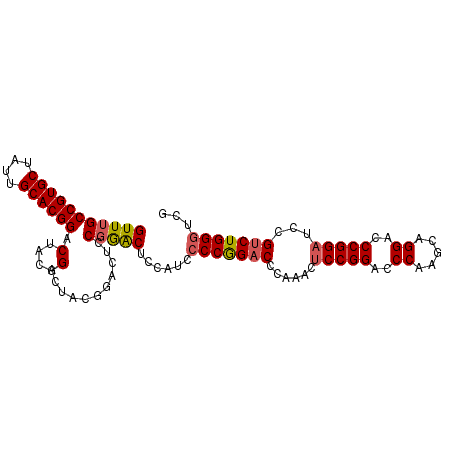
| Location | 12,909,428 – 12,909,527 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -28.73 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12909428 99 - 22407834 GUUUGCCGUGCUAUUGCACGGACUACGGACUACGGACUCCGGACUCCAUCCCCGGACCCAAACUCCGGACCCAAGCAGGACCCGGAUCCGUCUGGGUCG (((((((((((....))))))....((((....((..(((((.........))))).))....))))....)))))..((((((((....)))))))). ( -42.50) >DroSec_CAF1 87081 85 - 1 GUUUGCCGUGCUAUUGCACGG--------------ACUCCGGACACCAUCCCCGGACCCAAACUCCGGACCCAAGUAGGACCCGGAUCCGUCUGGGUCG .....((((((....))))))--------------....(((.........)))((((((.(((((((..((.....))..)))))...)).)))))). ( -35.00) >DroSim_CAF1 92155 99 - 1 GUUUGCCGUGCUAUUGCACGGACUACGGACUACGGACUCCGGACCCCAUCCCCAGACCCAAACCCCGGACCCAAGCAGGACCCGGAUCCGUCUGGGUCG ((...((((((....))))))...))((((....)..)))(((.....)))...((((((.((.((((..((.....))..))))....)).)))))). ( -36.90) >DroEre_CAF1 92246 92 - 1 GUUUGCCGUGCUAUUGCACGGACUACGGACUCCAG--U-CCAGCUCCAUCCCCGGACCCGAACUCCGGAUCCAAGCAGGACCCGGAUCCGGCUGG---- ((...((((((....))))))...))((((....)--)-))..........((((.((.((..(((((.(((.....))).))))))).))))))---- ( -38.90) >consensus GUUUGCCGUGCUAUUGCACGGACUACGGACUACGGACUCCGGACUCCAUCCCCGGACCCAAACUCCGGACCCAAGCAGGACCCGGAUCCGUCUGGGUCG (((((((((((....)))))).(....)...........)))))......(((((((......(((((..((.....))..)))))...)))))))... (-28.73 = -29.22 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:21 2006