
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,903,756 – 12,903,897 |
| Length | 141 |
| Max. P | 0.819570 |

| Location | 12,903,756 – 12,903,874 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12903756 118 + 22407834 GCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA-AUUUCUGCACGCAAUU ..........(((((((((.(((((((.....(((...((...((....))...((((((((...(((((((...)))))))))))))))..))..)))..)-))))))))).)))))) ( -33.20) >DroSec_CAF1 81383 118 + 1 GCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA-AUUUCUGCACGCAAUU ..........(((((((((.(((((((.....(((...((...((....))...((((((((...(((((((...)))))))))))))))..))..)))..)-))))))))).)))))) ( -33.20) >DroSim_CAF1 86077 118 + 1 GCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA-AUUUCUGCACGCAAUU ..........(((((((((.(((((((.....(((...((...((....))...((((((((...(((((((...)))))))))))))))..))..)))..)-))))))))).)))))) ( -33.20) >DroEre_CAF1 86791 114 + 1 GCCAAAAAAAC--UG--GCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA-AUUCCUGCACGCAAUG ((((.......--))--)).....((((((........................((((((((...(((((((...))))))))))))))).(((((......-...))))).)))))). ( -35.10) >DroYak_CAF1 83765 119 + 1 GCCAAAAAAAGAUUGCUCCUGGAAAUUGGGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAAUUCCUGCACGCAAUU ..........(((((((((..(...)..)))............((.........((((((((...(((((((...))))))))))))))).(((((..........))))))))))))) ( -31.70) >DroAna_CAF1 102957 111 + 1 ------AAAGCACUGCAACUGGAAGUGGGCUAAGGGAAUAAGCACGAGAAAUAAAUUUGGCAUAAGCACCUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA-AA-UCUGCACGCAAUU ------..(((.(..(........)..))))..........((.............((((((...((((.........))))))))))...(((((......-..-)))))..)).... ( -24.50) >consensus GCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAA_AUUUCUGCACGCAAUU .......................(((((((........................(((((((...((((((((...))))))))))))))).(((((..........))))).))))))) (-24.72 = -25.38 + 0.67)



| Location | 12,903,795 – 12,903,897 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12903795 102 + 22407834 GAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUUCUGCACGCAAUUCUUCUCU--CGUCCUUUUGAAAAAA ((((((((.......((((((((...(((((((...))))))))))))))).(((((.........)))))..............)--)).)))))........ ( -26.50) >DroPse_CAF1 96439 97 + 1 GCGAAUGAAAA-----UUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAAUUUCUGCACGCCAUUCUUCUCGAAGGUCCUUUAG-A-AAA ..(((((....-----.((((((...(((((((...)))))))))))))...(((((.........)))))....)))))((((.((((...))))))-)-).. ( -25.20) >DroSec_CAF1 81422 101 + 1 GAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUUCUGCACGCAAUUCUUCCCU--CGUCCUUUUGGA-AAA ((.((.(((......((((((((...(((((((...))))))))))))))).(((((.........)))))......)))..)).)--).(((....)))-... ( -28.00) >DroEre_CAF1 86826 101 + 1 GAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUCCUGCACGCAAUGCUGCUCU--CGUCCUUUUGCG-AAA ((((((((.......((((((((...(((((((...))))))))))))))).(((((.........)))))..(((....)))..)--)).)))))....-... ( -30.10) >DroAna_CAF1 102990 99 + 1 AAGCACGAGAAAUAAAUUUGGCAUAAGCACCUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAA-UCUGCACGCAAUUCUUCUCU--AGUCCUUUGG-A-GAA ..((.............((((((...((((.........))))))))))...(((((........-)))))..))......(((((--((....))))-)-)). ( -25.30) >DroPer_CAF1 116524 97 + 1 GCGAAUGAAAA-----UUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAAUUUCUGCACGCCAUUCUUCUCGAAGGUCCUUUAG-A-AAA ..(((((....-----.((((((...(((((((...)))))))))))))...(((((.........)))))....)))))((((.((((...))))))-)-).. ( -25.20) >consensus GAAGAUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUUCUGCACGCAAUUCUUCUCU__CGUCCUUUUG_A_AAA .................(((((...((((((((...)))))))))))))...(((((.........)))))................................. (-18.81 = -18.70 + -0.11)

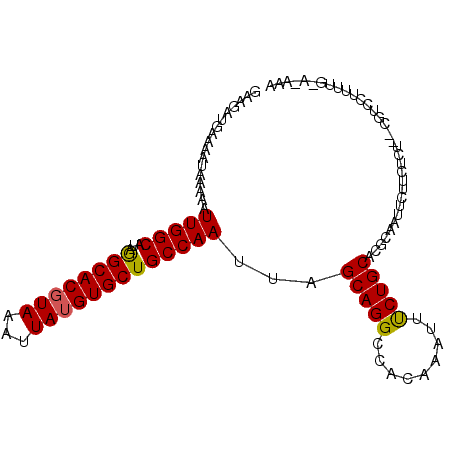
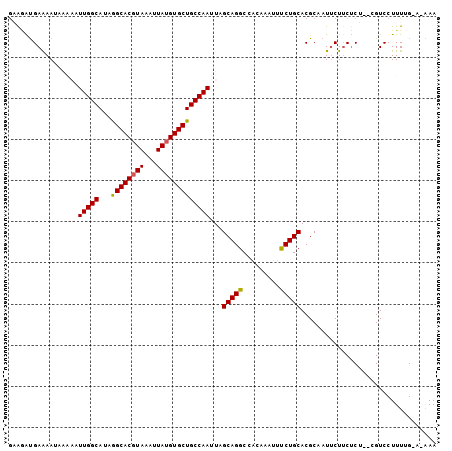
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:19 2006