
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,899,639 – 12,899,731 |
| Length | 92 |
| Max. P | 0.826774 |

| Location | 12,899,639 – 12,899,731 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.61 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
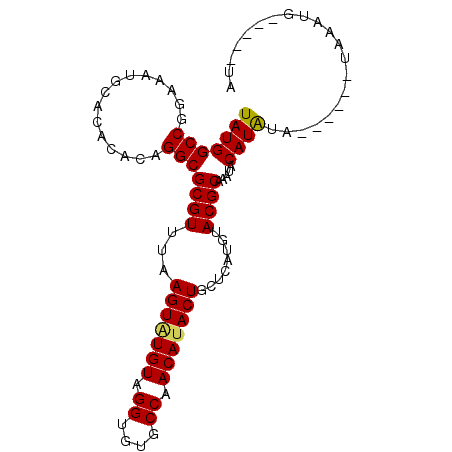
>2L_DroMel_CAF1 12899639 92 + 22407834 UAUGGCCGGAAAUGCACACACUGGCGCGUUUAAGUAUGUAGGUGUGCCAACAUACUGCUCAUGUACGCAAAUACAUAUA------UGUAUGUAUUGUA ....(((((...........)))))((((...(((((((.((....)).)))))))........)))).((((((((..------..))))))))... ( -27.00) >DroSec_CAF1 77261 86 + 1 UAUGGCCGGAAAUGCACACACAGGCGCGUUUAAGUAUGUAGGUGUGCCAACA-ACUGCUCAUGUACGCAAAUACAUAUA------UAAAUG-----UA ....(((...............)))(((((((.(((((((.((((((.....-.........))))))...))))))).------))))))-----). ( -22.60) >DroSim_CAF1 80317 87 + 1 UAUGGCCGGAAAUGCACACACAGGCGCGUUUAAGUAUGUAGGUGUGCCAACAUACUGCUCAUGUACGCAAAUACAUAUA------UGAAUG-----UA ....(((...............)))(((((((.(((((((.((((((...............))))))...))))))).------))))))-----). ( -22.62) >DroEre_CAF1 82723 93 + 1 UAUGGCCGGAAAUGCACACACAGGCGCGUUUAAGUGUGUAGGUGGGCCAACAUACUGCUCAUGUACGCAAAUACAUGUACUCGUACAUAUG-----UA ..(((((.(...(((((((..(((....)))..)))))))..).)))))(((((.((..(..(((((........)))))..)..))))))-----). ( -26.50) >consensus UAUGGCCGGAAAUGCACACACAGGCGCGUUUAAGUAUGUAGGUGUGCCAACAUACUGCUCAUGUACGCAAAUACAUAUA______UAAAUG_____UA (((((((...............)))((((...(((((((.((....)).)))))))........)))).....))))..................... (-19.67 = -19.61 + -0.06)

| Location | 12,899,639 – 12,899,731 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
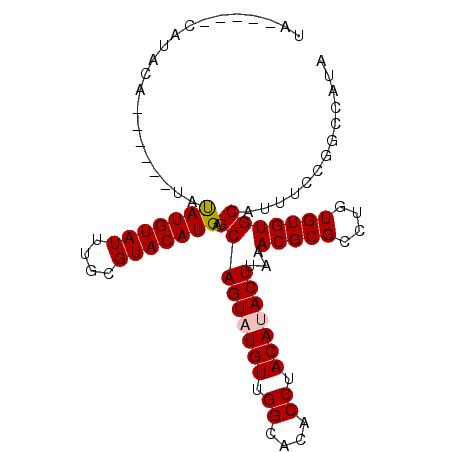
>2L_DroMel_CAF1 12899639 92 - 22407834 UACAAUACAUACA------UAUAUGUAUUUGCGUACAUGAGCAGUAUGUUGGCACACCUACAUACUUAAACGCGCCAGUGUGUGCAUUUCCGGCCAUA ...((((((((..------..))))))))(((((((((..(((((((((.((....)).))))))).......))..)))))))))............ ( -23.11) >DroSec_CAF1 77261 86 - 1 UA-----CAUUUA------UAUAUGUAUUUGCGUACAUGAGCAGU-UGUUGGCACACCUACAUACUUAAACGCGCCUGUGUGUGCAUUUCCGGCCAUA ((-----(((...------...)))))..(((((((((..(((((-(((.((....)).))).))).......))..)))))))))............ ( -16.11) >DroSim_CAF1 80317 87 - 1 UA-----CAUUCA------UAUAUGUAUUUGCGUACAUGAGCAGUAUGUUGGCACACCUACAUACUUAAACGCGCCUGUGUGUGCAUUUCCGGCCAUA ((-----(((...------...)))))..(((((((((..(((((((((.((....)).))))))).......))..)))))))))............ ( -21.41) >DroEre_CAF1 82723 93 - 1 UA-----CAUAUGUACGAGUACAUGUAUUUGCGUACAUGAGCAGUAUGUUGGCCCACCUACACACUUAAACGCGCCUGUGUGUGCAUUUCCGGCCAUA ..-----...((((((..((.(((((((....))))))).)).))))))(((((....(((((((............))))))).......))))).. ( -25.60) >consensus UA_____CAUACA______UAUAUGUAUUUGCGUACAUGAGCAGUAUGUUGGCACACCUACAUACUUAAACGCGCCUGUGUGUGCAUUUCCGGCCAUA .....................(((((((....))))))).(((((((((.((....)).)))))))...(((((....)))))))............. (-18.11 = -18.43 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:17 2006