| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,893,330 – 12,893,437 |
| Length | 107 |
| Max. P | 0.644958 |

| Location | 12,893,330 – 12,893,437 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -14.26 |
| Energy contribution | -14.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
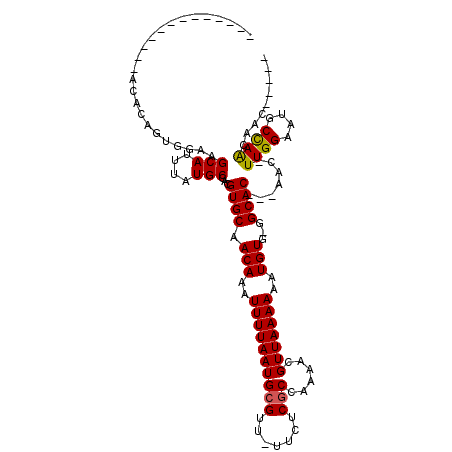
>2L_DroMel_CAF1 12893330 107 - 22407834 GGGCAGCACAC--ACACAGUGGAAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU-UUCUCGCCAAAACGUUAAAAAAUGUG-GGCAC---AAC-UUGGAAUGCCAACAAC----- .((((((.(((--.....)))...))...........((((.(((..(((((..(((((-((......))))))))))))..))).-.))))---...-......))))......----- ( -25.10) >DroPse_CAF1 85049 111 - 1 --GCA-CACUCGCACACAGUGGAAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU-UUCUCGCCAAAACGUUAAAAAAUGUG-GGCAC---AAC-UUGGAAUUCCAGCAACAACAA --(((-(..(((((...((((....))))..))))).))))............(((...-.....(((...(((((....))))).-)))..---...-.(((....))))))....... ( -28.50) >DroGri_CAF1 92983 101 - 1 -------------UCAAACACGCAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU--UCUCGCCAAAAUGUUAAAAAAUGUGCGGCAC---AAG-UUGGAAUGCCAAGAACAAAAA -------------.......((((.......))))..((((..((.(((((...(((((--(.......))))))...))))).))..))))---...-((((....))))......... ( -20.50) >DroWil_CAF1 101634 100 - 1 -----------AGAUAUAGUCAAAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUUUUUCUCACCAAAACGUUAAAAAAUGUG-AGCAC---AAGUUUGGAAUGCUAGCAAC----- -----------............(((((((..((...((((.(((..(((((..(((((((.......))))))))))))..))).-.))))---..))...)))))))......----- ( -22.40) >DroYak_CAF1 74859 112 - 1 GGGCAGCACACACACACAGUGGAAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU-UUUUCGCCAAAACGUUAAAAAAUGUG-GGCACAACAAC-UUGGAAUGCCAACAAC----- .((((...(((((.....)))...(((....)))...((((.(((..(((((..(((((-((......))))))))))))..))).-.))))......-.))...))))......----- ( -26.10) >DroAna_CAF1 94812 86 - 1 -----------------------AGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU-UUCUCGCCAAAACGUUAAAAAAUGUG-GGCAC---AAC-UUGGAAUGCCAACAAC----- -----------------------.((((((.......((((.(((..(((((..(((((-((......))))))))))))..))).-.))))---...-...)))))).......----- ( -19.62) >consensus _____________ACACAGUGGAAGCAUUUAUGCGACGUGCAACAAAUUUUAAUGCGUU_UUCUCGCCAAAACGUUAAAAAAUGUG_GGCAC___AAC_UUGGAAUGCCAACAAC_____ ........................(((....)))...((((.(((..((((((((((.......)))......)))))))..)))...)))).......((((....))))......... (-14.26 = -14.07 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:15 2006