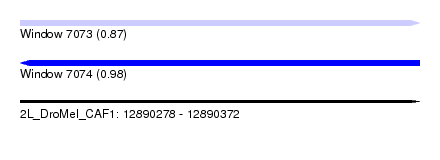
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,890,278 – 12,890,372 |
| Length | 94 |
| Max. P | 0.976405 |

| Location | 12,890,278 – 12,890,372 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
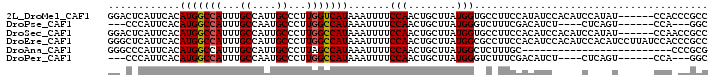
>2L_DroMel_CAF1 12890278 94 + 22407834 GGCGGGUGG------AUAUGGAUGUGGAUAUGGAAGGCACCAUAAGCAGUUGGAAAAUUUAUGACCAAGGGCAAUGGCAAAUGGCCAUGUGAAUGAGUCC (((..((..------((((((.(((.(.(((((......))))).((..((((...........))))..))..).))).....))))))..))..))). ( -20.90) >DroPse_CAF1 81819 84 + 1 GCC---UGG------ACUGAG----AGAUGUCGAAAGACCCAUAAGCAGUUGGAAAAUUUAUGGCCAAGGGCAUUGGCAAAUGGCCAUGUGAAUGGG--- .((---(.(------((((..----....(((....))).......))))).)......((((((((...((....))...)))))))).....)).--- ( -26.12) >DroSec_CAF1 69346 94 + 1 GGCGGUUGG------AUAUGGAUGUGGAUGUGGAAGGCACCAUAAGCAGUUGGAAAAUUUAUGGCCAAGGGCAAUGGCAAAUGGCCAUGUGAAUGAGUCC (((..((.(------((.((..(((((.(((.....))))))))..))))).)).....((((((((...((....))...)))))))).......))). ( -23.80) >DroEre_CAF1 74919 100 + 1 GGCGGGUGGAUAAGGAUGUGGAUGUGGAUGUGGAAGGCGCCAUAAGCAGUUGGAAAAUUUAUGGCCAAGGGCAAUGGCAAAUGGCCAUGUGAAUGAGCCC ...((((..((.....(((...(((((.(((.....)))))))).)))...........((((((((...((....))...))))))))...))..)))) ( -26.80) >DroAna_CAF1 92185 75 + 1 CGCGGG-------------------------GCAAAGAGCCAUAAGCAGUUGGAAAAUUUAUGGCUAAGGGCAAUGGCAAAUGGCCAUGUGAAUGGGCCC ....((-------------------------((....((((((((...(((....)))))))))))....(((.((((.....)))))))......)))) ( -24.40) >DroPer_CAF1 101654 84 + 1 GCC---UGG------ACUGAG----AGAUGUCGAAAGACCCAUAAGCAGUUGGAAAAUUUAUGGCCAAGGGCAUUGGCAAAUGGCCAUGUGAAUGGG--- .((---(.(------((((..----....(((....))).......))))).)......((((((((...((....))...)))))))).....)).--- ( -26.12) >consensus GGCGGGUGG______AUAUGG____GGAUGUGGAAAGAACCAUAAGCAGUUGGAAAAUUUAUGGCCAAGGGCAAUGGCAAAUGGCCAUGUGAAUGAG_CC .......................................(((........)))......((((((((...((....))...))))))))........... (-15.01 = -15.03 + 0.03)

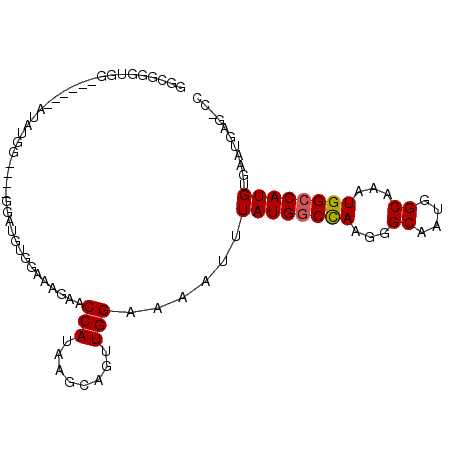
| Location | 12,890,278 – 12,890,372 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12890278 94 - 22407834 GGACUCAUUCACAUGGCCAUUUGCCAUUGCCCUUGGUCAUAAAUUUUCCAACUGCUUAUGGUGCCUUCCAUAUCCACAUCCAUAU------CCACCCGCC (((.........(((((((...((....))...)))))))............((..(((((......)))))..))..)))....------......... ( -18.10) >DroPse_CAF1 81819 84 - 1 ---CCCAUUCACAUGGCCAUUUGCCAAUGCCCUUGGCCAUAAAUUUUCCAACUGCUUAUGGGUCUUUCGACAUCU----CUCAGU------CCA---GGC ---.((......(((((((...((....))...)))))))..........((((.......(((....)))....----..))))------...---)). ( -18.22) >DroSec_CAF1 69346 94 - 1 GGACUCAUUCACAUGGCCAUUUGCCAUUGCCCUUGGCCAUAAAUUUUCCAACUGCUUAUGGUGCCUUCCACAUCCACAUCCAUAU------CCAACCGCC ((..........(((((((...((....))...)))))))............((..(((((((.............)).))))).------.)).))... ( -18.72) >DroEre_CAF1 74919 100 - 1 GGGCUCAUUCACAUGGCCAUUUGCCAUUGCCCUUGGCCAUAAAUUUUCCAACUGCUUAUGGCGCCUUCCACAUCCACAUCCACAUCCUUAUCCACCCGCC ((((........(((((((...((....))...))))))).............((.....)))))).................................. ( -19.00) >DroAna_CAF1 92185 75 - 1 GGGCCCAUUCACAUGGCCAUUUGCCAUUGCCCUUAGCCAUAAAUUUUCCAACUGCUUAUGGCUCUUUGC-------------------------CCCGCG ((((........(((((.....))))).......((((((((.............))))))))....))-------------------------)).... ( -19.72) >DroPer_CAF1 101654 84 - 1 ---CCCAUUCACAUGGCCAUUUGCCAAUGCCCUUGGCCAUAAAUUUUCCAACUGCUUAUGGGUCUUUCGACAUCU----CUCAGU------CCA---GGC ---.((......(((((((...((....))...)))))))..........((((.......(((....)))....----..))))------...---)). ( -18.22) >consensus GG_CCCAUUCACAUGGCCAUUUGCCAUUGCCCUUGGCCAUAAAUUUUCCAACUGCUUAUGGCGCCUUCCACAUCC____CCAAAU______CCACCCGCC ............(((((((...((....))...))))))).......(((........)))....................................... (-11.99 = -12.02 + 0.03)
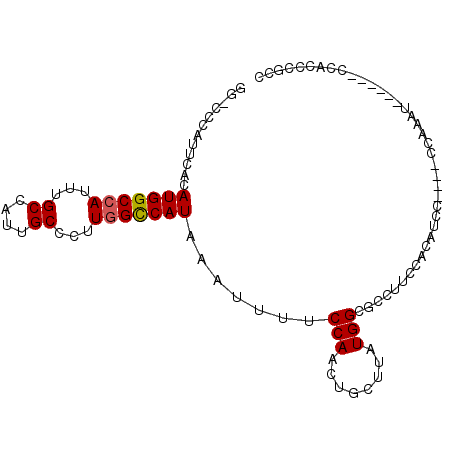
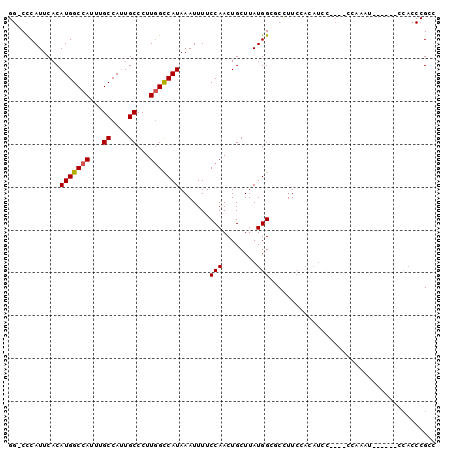
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:14 2006