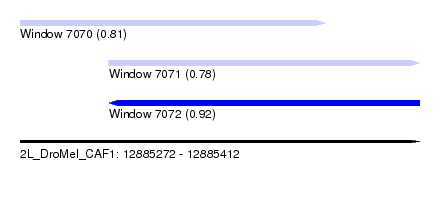
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,885,272 – 12,885,412 |
| Length | 140 |
| Max. P | 0.919741 |

| Location | 12,885,272 – 12,885,379 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12885272 107 + 22407834 UUUGUUUGUCUCACUAUCAGACGAACAUCCGCUGACGCCGAGACUUCCUCUUCCUCUUCGCCCGUGUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((....((.((((...........)))).))..(((..((...............))...)))....)))))..... ( -23.36) >DroSec_CAF1 64419 102 + 1 UUUGUUUGUCUCACUAUCAGACGAACAUCCGCUGACGCCGGGACUUCCUCUAC-----CGCCCGCAUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((((..(((((((..........-----.......................)))))))..)).....)))))..... ( -26.61) >DroSim_CAF1 67224 102 + 1 UUUGUUUGUCUCACUAUCAGACGAACAUCCGCUGACGCCGAGACUUCCUCUAC-----CGCCCGCAUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((.......(((.....)))...-----............................(((.....))))))))..... ( -21.70) >DroEre_CAF1 70139 95 + 1 UUUGUUUGUCUCACUAUCAGACGAACAUCCGCUGACGCCGAGGCUUCCUCCAC-----CGCCCG-------UAUUCCUUGUUGCCGGUCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((.......((((...))))...-----.(((.(-------((........))).))).........)))))..... ( -25.60) >DroYak_CAF1 66561 95 + 1 UUUGUUUGUCUCACUAUCAGACGAGCAUCCGCUGACGCCGAGACUUCCUCUAC-----CGCCCA-------UAUUCCCUAUUCCCGGGCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((.......(((.....)))...-----.((((.-------..............))))........)))))..... ( -25.66) >consensus UUUGUUUGUCUCACUAUCAGACGAACAUCCGCUGACGCCGAGACUUCCUCUAC_____CGCCCG__UUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGU ..(((((((((.......)))))))))(((((.......(((.....)))....................................(((.....))))))))..... (-20.92 = -20.80 + -0.12)

| Location | 12,885,303 – 12,885,412 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12885303 109 + 22407834 CUGACGCCGAGACUUCCUCUUCCUCUUCGCCCGUGUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGUAUCCUCGUCUAUUCCCAAGCUGCAUUUGCAGCU ..((((..(((.....)))...........(((((((.......((........)).......)))))))...........))))........((((((....)))))) ( -25.84) >DroSec_CAF1 64450 104 + 1 CUGACGCCGGGACUUCCUCUAC-----CGCCCGCAUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGUAUCCUCGUCUAUUCCCAAGCUGCAUUUGCAGCU .((..(((((((..........-----.......................)))))))..)).(((((.(((......))).))))).......((((((....)))))) ( -29.31) >DroSim_CAF1 67255 104 + 1 CUGACGCCGAGACUUCCUCUAC-----CGCCCGCAUUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGUAUCCUCGUCUAUUCCCAAGCUGCAUUUGCAGCU .((..((((((.....)))...-----.(....)....................)))..)).(((((.(((......))).))))).......((((((....)))))) ( -23.60) >DroEre_CAF1 70170 97 + 1 CUGACGCCGAGGCUUCCUCCAC-----CGCCCG-------UAUUCCUUGUUGCCGGUCCCACAGACGCGGAACUGUAUCCUCAACUAUUCCCAGGCUGCAUUUGCAGCU (((..((.((((...))))...-----.(((.(-------((........))).))).........))((((..((.......))..)))))))(((((....))))). ( -23.70) >DroYak_CAF1 66592 97 + 1 CUGACGCCGAGACUUCCUCUAC-----CGCCCA-------UAUUCCCUAUUCCCGGGCCCACAGACGCGGAACUGUAUCCUCGACUAUUCCCAAGCUGCAUUUGCAGCU (((.....(((.....)))...-----.((((.-------..............))))...))).((.(((......))).))..........((((((....)))))) ( -23.56) >consensus CUGACGCCGAGACUUCCUCUAC_____CGCCCG__UUCCUUAUUCCUUAUUCCCGGUCCCACAGACGCGGAACUGUAUCCUCGUCUAUUCCCAAGCUGCAUUUGCAGCU ........(((.....)))...................................((......(((((.(((......))).)))))....)).((((((....)))))) (-19.22 = -19.50 + 0.28)


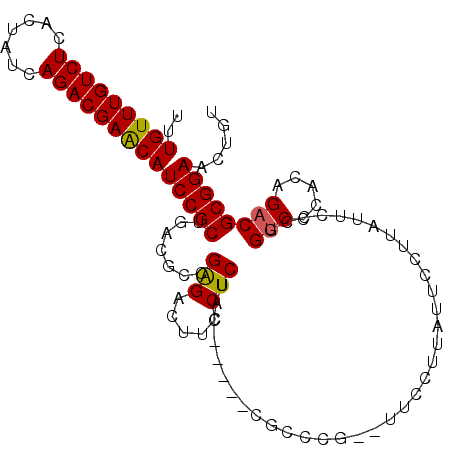
| Location | 12,885,303 – 12,885,412 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12885303 109 - 22407834 AGCUGCAAAUGCAGCUUGGGAAUAGACGAGGAUACAGUUCCGCGUCUGUGGGACCGGGAAUAAGGAAUAAGGAACACGGGCGAAGAGGAAGAGGAAGUCUCGGCGUCAG ((((((....))))))........((((.((((.(..((((.((((((((...((...............))..))))))))....))))..)...))))...)))).. ( -34.86) >DroSec_CAF1 64450 104 - 1 AGCUGCAAAUGCAGCUUGGGAAUAGACGAGGAUACAGUUCCGCGUCUGUGGGACCGGGAAUAAGGAAUAAGGAAUGCGGGCG-----GUAGAGGAAGUCCCGGCGUCAG ((((((....)))))).....(((((((.(((......))).)))))))((..((((((...............(((.....-----))).......))))))..)).. ( -33.05) >DroSim_CAF1 67255 104 - 1 AGCUGCAAAUGCAGCUUGGGAAUAGACGAGGAUACAGUUCCGCGUCUGUGGGACCGGGAAUAAGGAAUAAGGAAUGCGGGCG-----GUAGAGGAAGUCUCGGCGUCAG .(((((....)))))((((..(((((((.(((......))).)))))))....)))).....................((((-----...(((.....)))..)))).. ( -32.70) >DroEre_CAF1 70170 97 - 1 AGCUGCAAAUGCAGCCUGGGAAUAGUUGAGGAUACAGUUCCGCGUCUGUGGGACCGGCAACAAGGAAUA-------CGGGCG-----GUGGAGGAAGCCUCGGCGUCAG .(((((....)))))....((...(((((((......(((((((((((((...((........))..))-------))))).-----))))))....))))))).)).. ( -37.70) >DroYak_CAF1 66592 97 - 1 AGCUGCAAAUGCAGCUUGGGAAUAGUCGAGGAUACAGUUCCGCGUCUGUGGGCCCGGGAAUAGGGAAUA-------UGGGCG-----GUAGAGGAAGUCUCGGCGUCAG ((((((....))))))...((...(((((((......((((.((((((((..(((.......)))..))-------))))))-----.....)))).))))))).)).. ( -36.90) >consensus AGCUGCAAAUGCAGCUUGGGAAUAGACGAGGAUACAGUUCCGCGUCUGUGGGACCGGGAAUAAGGAAUAAGGAA__CGGGCG_____GUAGAGGAAGUCUCGGCGUCAG .(((((....)))))((((..(((((((.(((......))).)))))))....))))...................(((((...............))).))....... (-26.76 = -26.52 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:12 2006