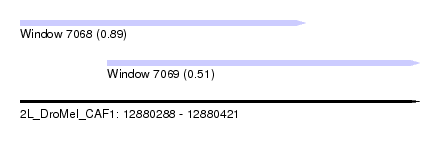
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,880,288 – 12,880,421 |
| Length | 133 |
| Max. P | 0.885925 |

| Location | 12,880,288 – 12,880,383 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12880288 95 + 22407834 ---------UCCCAUCCCCUACUGGCUCUUUUGCUGCGCUGGAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU ---------..(((........))).(((..((((((..((((....)))).)))).))..))).((((((((...((((......)))).))))))))..... ( -23.10) >DroSec_CAF1 59395 94 + 1 ---------UCCCAUC-CCUACUGGCUCUUUUGCUGCGCUCAAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU ---------.......-.(((.(((((.....(((................)))))))).)))..((((((((...((((......)))).))))))))..... ( -20.09) >DroSim_CAF1 62314 94 + 1 ---------UCCCAUC-CCUACUGGCUCUUUUGCUGCGCUCAAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU ---------.......-.(((.(((((.....(((................)))))))).)))..((((((((...((((......)))).))))))))..... ( -20.09) >DroEre_CAF1 65250 81 + 1 ---------CCCC--------------CUCUUGCUGCGCGGCAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU ---------....--------------.(((.(((((..(....).......)))))....))).((((((((...((((......)))).))))))))..... ( -18.80) >DroYak_CAF1 61585 94 + 1 ---------CCCCAUU-CCUGCUGGCUCUUUUGCUUCGCGGGAACUUUUCAAGCAGCUAUUACAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACUAUU ---------.....((-(((((.(((......)))..))))))).....................((((((((...((((......)))).))))))))..... ( -24.10) >DroAna_CAF1 81131 95 + 1 CACCCUUAACCCC----ACCACUUGUUCUUUUUCU-----GAAACUUUGCAAGCAGCCAUUAAAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU .............----....(((((.........-----........)))))............((((((((...((((......)))).))))))))..... ( -12.93) >consensus _________CCCCAUC_CCUACUGGCUCUUUUGCUGCGCUGAAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUU ................................(((((...............)))))........((((((((...((((......)))).))))))))..... (-14.18 = -14.84 + 0.67)

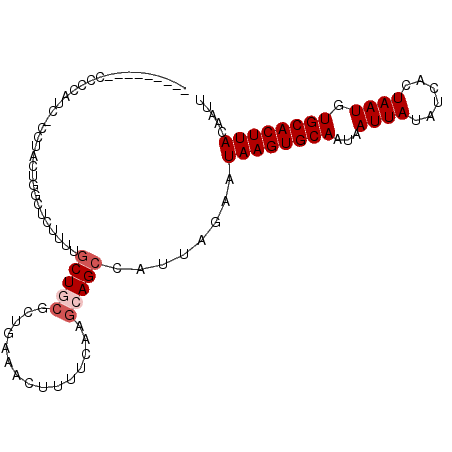
| Location | 12,880,317 – 12,880,421 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -15.85 |
| Energy contribution | -17.05 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12880317 104 + 22407834 CUGGAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUUGUUGACUUUUCGACUUGAUGGCAGUGAUGGCGCACCCC ..((.............((((((((..((((((((...((((......)))).)))))))).....(((((....))))))))))))).(((....))).)).. ( -26.90) >DroSim_CAF1 62342 104 + 1 CUCAAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUUGUUGACUUUUCGUCUUGAUGGCAGUGACGCAGCCCCCC ..........(((....((((((((..((((((((...((((......)))).))))))))........(((.....)))))))))))..)))........... ( -23.00) >DroEre_CAF1 65265 102 + 1 CGGCAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUUGUUGACUUUUCCUCUUGAUGGCAGGAACGCAACCCC-- .(((..((.....))..))).......((((((((...((((......)))).)))))))).....((((....(((((((....)).)))))..))))...-- ( -23.10) >DroYak_CAF1 61613 102 + 1 CGGGAACUUUUCAAGCAGCUAUUACAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACUAUUGUUGACUUUUCGUCUUGCUGGCAGGGACGCAGCCCC-- .(((.....(((..(((((....((((((((((((...((((......)))).))))))...)))))).(((.....)))..))).))..))).....))).-- ( -24.90) >DroAna_CAF1 81162 101 + 1 --GAAACUUUGCAAGCAGCCAUUAAAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUUGUUGACUUUUCUUCUUAAUAGCGCGACUACACCCCU-C --......(((((((((((.(((....((((((((...((((......)))).)))))))).))).)))).))).....((....)).))))..........-. ( -15.80) >DroPer_CAF1 90957 101 + 1 ---CAACUCUGCCAGAGGCCAUCAGAUUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUCACUUUUGUUGACUGUUCGUCUUGAUAAGAGGGCCAGGGCCCCGC ---.......(((...((((.........((((((...((((......)))).))))))..(((((((((((.....)))..))))))))))))..)))..... ( -29.00) >consensus C_GAAACUUUUCAAGCAGCCAUUAGAAUAAGUGCAAUAAUUAUAUCACUAAUGUGCACUUACAAUUGUUGACUUUUCGUCUUGAUGGCAGGGACGCAGCCCC_C .................((((((((..((((((((...((((......)))).))))))))........(((.....)))))))))))................ (-15.85 = -17.05 + 1.20)

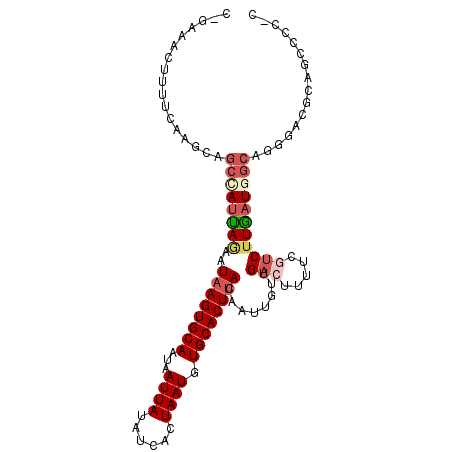
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:10 2006