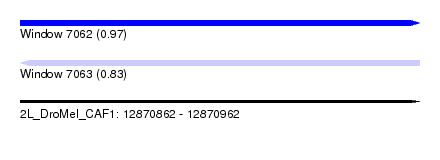
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,870,862 – 12,870,962 |
| Length | 100 |
| Max. P | 0.968839 |

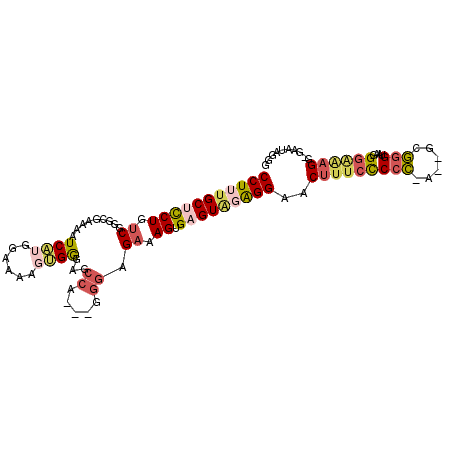
| Location | 12,870,862 – 12,870,962 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -12.94 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12870862 100 + 22407834 CCCUAUUC-CCUUUCCCUUACCCUCACU-GGGAGAAAGUUCCUCUACUCACUUUCUCCC---UGGCUCCCACUUUUCCAUGAUUUUCGCCCGACAGGGGCAAAGG ........-((((((((((.........-((((((((((..........))))))))))---.(((.....................)))....))))).))))) ( -27.30) >DroSec_CAF1 49956 100 + 1 CCCUAUUC-CCUUUCCAUUACCCGCUGU-GGGGGCAAGUUCCUCUACUCACUUUCUCCA---UGGCUCCCACUUUUCCAUGAUUUUCGCCCGACAGGAGCAAAGG ........-(((((.........(((((-(((((.((((..........))))))))))---)))).....................((((....)).))))))) ( -22.50) >DroSim_CAF1 52548 100 + 1 CCCUAUUC-CCUUUCCAUUACCCGCUCU-GGGGGAAAGUUCCUCUACUCACUUUCUCCC---UGGCUCCCACUUUUCCAUGAUUUUCGCCCGACAGGAGCAAAGG ........-(((((.........(((..-((((((((((..........))))))))))---.))).....................((((....)).))))))) ( -24.80) >DroEre_CAF1 55939 97 + 1 CCCCUUUG-CCUGUCCCUCCCCCGC----GGGGGAAAGUUCCCACACUCACUUUCUCCC---GGGCACCCGCUUUUCCAUGAUUUUAGUCCGACAGGAGCAAAGG ..((((((-((((((........((----((((((((((..........))))))))))---((....))))........(((....))).)))))..))))))) ( -34.30) >DroYak_CAF1 52105 100 + 1 CCCCUUUUCCCUUUCCCACCCCCCC--UGGGGGGAAAGUUCCUACACUCACUUUCUCCG---GGGCACCCACUUUUCCUUGAUUUUAGCCCGACAGGAGCAAAGG ..........(((((((.(((....--.))))))))))............((((((((.---((((.....................))))....)))).)))). ( -29.20) >DroAna_CAF1 73017 97 + 1 CUCUACCC-CCACCUCGUUGCGAUC----CGGGGAAAGUUCCUCAACUCACUUUCUCCUCCCGCCCGGUCGGUCCUCCACGAUUC---CCCGCCAGAUGCAAAGG ........-...(((..((((.(((----.(((((((((..........))))))))).......(((..((((......)))).---.)))...)))))))))) ( -22.50) >consensus CCCUAUUC_CCUUUCCCUUACCCGC__U_GGGGGAAAGUUCCUCUACUCACUUUCUCCC___GGGCUCCCACUUUUCCAUGAUUUUCGCCCGACAGGAGCAAAGG .........(((((................(((((((((..........)))))))))......(((((..........................)))))))))) (-12.94 = -13.94 + 1.00)

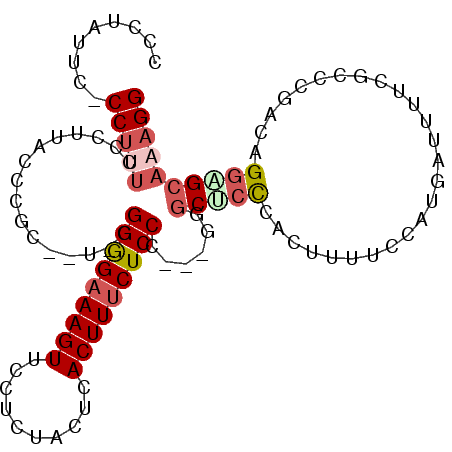
| Location | 12,870,862 – 12,870,962 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12870862 100 - 22407834 CCUUUGCCCCUGUCGGGCGAAAAUCAUGGAAAAGUGGGAGCCA---GGGAGAAAGUGAGUAGAGGAACUUUCUCCC-AGUGAGGGUAAGGGAAAGG-GAAUAGGG (((((((((......(((.....((((......))))..))).---((((((((((..........))))))))))-.....))))))))).....-........ ( -37.50) >DroSec_CAF1 49956 100 - 1 CCUUUGCUCCUGUCGGGCGAAAAUCAUGGAAAAGUGGGAGCCA---UGGAGAAAGUGAGUAGAGGAACUUGCCCCC-ACAGCGGGUAAUGGAAAGG-GAAUAGGG ((.((((((((((.(((......((((((...........)))---))).....(..(((......)))..)))).-)))).)))))).)).....-........ ( -28.50) >DroSim_CAF1 52548 100 - 1 CCUUUGCUCCUGUCGGGCGAAAAUCAUGGAAAAGUGGGAGCCA---GGGAGAAAGUGAGUAGAGGAACUUUCCCCC-AGAGCGGGUAAUGGAAAGG-GAAUAGGG (((((((((((.((.(((.....((((......))))..))).---....)).)).)))))))))..(((((((((-.....)))....)))))).-........ ( -32.10) >DroEre_CAF1 55939 97 - 1 CCUUUGCUCCUGUCGGACUAAAAUCAUGGAAAAGCGGGUGCCC---GGGAGAAAGUGAGUGUGGGAACUUUCCCCC----GCGGGGGAGGGACAGG-CAAAGGGG (((((((..(((((...(((...((((.......(((....))---).......))))...)))...((((((((.----..))))))))))))))-)))))).. ( -38.74) >DroYak_CAF1 52105 100 - 1 CCUUUGCUCCUGUCGGGCUAAAAUCAAGGAAAAGUGGGUGCCC---CGGAGAAAGUGAGUGUAGGAACUUUCCCCCCA--GGGGGGGUGGGAAAGGGAAAAGGGG (((((((((((.(((((.((...(((........))).)).))---)))....)).)))).......((((((((((.--....))).)))))))...))))).. ( -34.30) >DroAna_CAF1 73017 97 - 1 CCUUUGCAUCUGGCGGG---GAAUCGUGGAGGACCGACCGGGCGGGAGGAGAAAGUGAGUUGAGGAACUUUCCCCG----GAUCGCAACGAGGUGG-GGGUAGAG .((((((.(((.((((.---...))))...)))((.(((..((((..((.((((((..........)))))).)).----..)))).....))).)-).)))))) ( -31.40) >consensus CCUUUGCUCCUGUCGGGCGAAAAUCAUGGAAAAGUGGGAGCCA___GGGAGAAAGUGAGUAGAGGAACUUUCCCCC_A__GCGGGUAAGGGAAAGG_GAAUAGGG (((((((((((.((.........((((......))))...((.....)).)).)).)))))))))..(((((((((......)))....)))))).......... (-16.35 = -17.58 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:04 2006