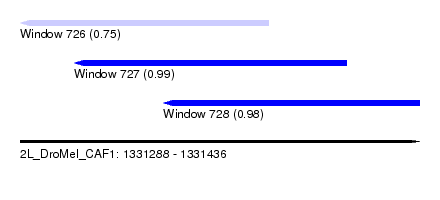
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,331,288 – 1,331,436 |
| Length | 148 |
| Max. P | 0.993167 |

| Location | 1,331,288 – 1,331,380 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -11.61 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
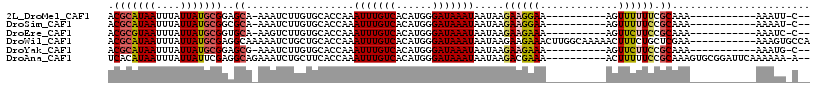
>2L_DroMel_CAF1 1331288 92 - 22407834 ACGCAUAAUUUAUUAUGCGGAGCA-AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGGAA----------AGUUUUUUCGCAAA-----------AAAUU-C-- .(((((((....)))))))..((.-..((((..((...((....))...))..)))).........((((((.----------...))))))))...-----------.....-.-- ( -18.20) >DroSim_CAF1 92178 92 - 1 ACGCAUAAUUUAUUAUGCGGCGCA-AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGGAA----------AGUUUUUCCGCAAA-----------AAAAU-C-- .(((((((....)))))))(((((-(....)))))).....(((((((......)))))))........((((----------.....)))).....-----------.....-.-- ( -22.00) >DroEre_CAF1 101877 92 - 1 ACGCGUAAUUUAUUAUGCGGUGCA-AAGUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA----------AGUUCUUCCGCAAA-----------AAAUC-C-- ..((((((....))))))((((((-........))))))..(((((((......))))))).....(((((..----------...)))))......-----------.....-.-- ( -22.60) >DroWil_CAF1 124848 106 - 1 ACGCAUAAUUUAUUAUGCGAGGCAAAAAUCUGCUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAACUUGGCAAAAACUUUCUGCUCGAA-----------AAAGUGCCA .(((((((....))))))).((((..........((.....(((((((......))))))).......(((((.((......)).))))))).....-----------....)))). ( -22.31) >DroYak_CAF1 92214 92 - 1 ACGCAUAAUUUAUUAUGCGGAGCG-AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA----------AGUUCUUCCGCAAA-----------AAAUG-C-- .(((((((....)))))))..(((-..((((..((...((....))...))..)))).........(((((..----------...))))))))...-----------.....-.-- ( -21.20) >DroAna_CAF1 116448 104 - 1 UCACAUAAUUUAUUAUUCGAGGCAGAAAUCUGCUUCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGACGAAA----------ACUUUUUCCGCAAAGUGCGGAUUCAAAAAA-A-- ........(((((((...(((((((....)))))))......((((((......)))))))))))))......----------......(((((.....))))).........-.-- ( -24.70) >consensus ACGCAUAAUUUAUUAUGCGGAGCA_AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA__________AGUUUUUCCGCAAA___________AAAUU_C__ .(((((((....)))))))..((..................(((((((......))))))).....((((((.............)))))).))....................... (-11.61 = -11.92 + 0.31)
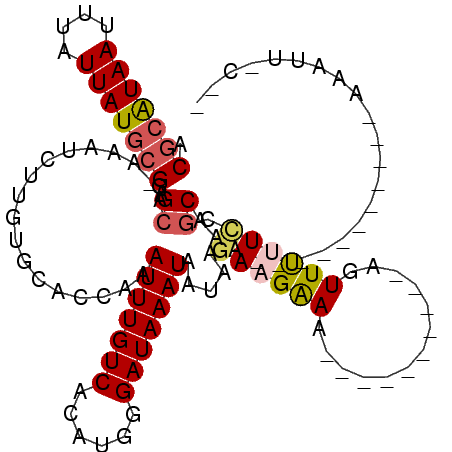
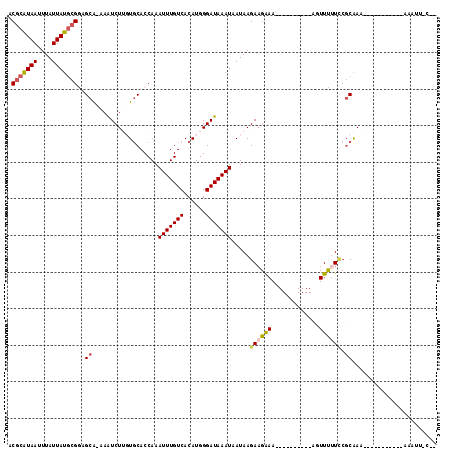
| Location | 1,331,308 – 1,331,409 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -13.39 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1331308 101 - 22407834 AGUUCC-U-UUUUUUCG--CCUUCCCAACUGCAACGCAUAAUUUAUUAUGCGGAGCA-AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGGAA----- ..((((-(-((((....--...(((((..(((..(((((((....)))))))..)))-......((((((.......)).)))))))))........)))))))))----- ( -30.56) >DroVir_CAF1 136064 105 - 1 UUUCUC-UUUCUAUUUUUU-----UACACUGCUACGCAUAAUUUAUUAUGCGAGGCAAAAAUCUGCUGCAGCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAACUUAC ....((-((..((((....-----....((((..(((((((....))))))).((((......))))))))...(((((((......)))))))))))..))))....... ( -24.50) >DroGri_CAF1 124472 107 - 1 AUUCUU-UUUUUUUUUG--CUUGU-GCAGUUUAACGCAUAAUUUAUUAUGCGAGGCAAAAAUCUGCUGCUGCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAACUUGC .(((((-(((.......--....(-(((((....(((((((....))))))).((((......)))))))))).(((((((......)))))))...))))))))...... ( -27.70) >DroEre_CAF1 101897 100 - 1 UCUAU--C-UUUUUUCG--CCUUCCCAGCUGCAACGCGUAAUUUAUUAUGCGGUGCA-AAGUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA----- ....(--(-(((((...--...(((((..(((((.((((((....))))))((((((-........))))))....))).))..)))))........)))))))..----- ( -29.04) >DroYak_CAF1 92234 101 - 1 AGUUUU-U-UUUUUUCG--CCCUCCCAACUGCAACGCAUAAUUUAUUAUGCGGAGCG-AAAUCUUGUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA----- ..((((-(-(((.....--...(((((..(((..(((((((....)))))))..)))-......((((((.......)).)))))))))........)))))))).----- ( -24.49) >DroAna_CAF1 116479 103 - 1 AGUUCCUU-UUUUUUGG--GGCACCCCACUGCGUCACAUAAUUUAUUAUUCGAGGCAGAAAUCUGCUUCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGACGAAA----- ........-.....(((--(....))))...((((................(((((((....))))))).....(((((((......))))))).....))))...----- ( -27.20) >consensus AGUUUC_U_UUUUUUCG__CCUUCCCAACUGCAACGCAUAAUUUAUUAUGCGAGGCA_AAAUCUGCUGCACCAAAUUUGUCACAUGGGAUAAAUAAUAAGAAGAAA_____ .........(((((((.............(((..(((((((....)))))))..))).................(((((((......))))))).....)))))))..... (-13.39 = -13.83 + 0.45)

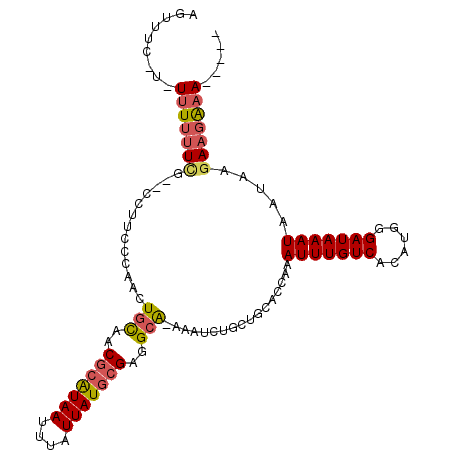
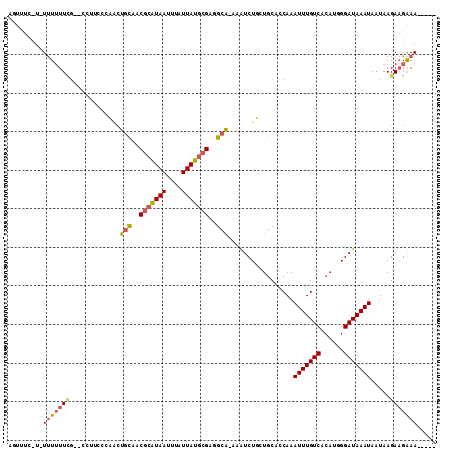
| Location | 1,331,341 – 1,331,436 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -15.76 |
| Consensus MFE | -9.57 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1331341 95 - 22407834 CAUUUUCUUU-CCCUAUAAUUUAAUUUCAGUUCC-U-UUUUUUCGCCUUCCCAACUGCAACGCAUAAUUUAUUAUGCGGAGCA-AAAUCUUGUGCACCA ..........-.......................-.-.......((.........(((..(((((((....)))))))..)))-.........)).... ( -13.27) >DroGri_CAF1 124510 95 - 1 UCCUUUUUU--ACAUAUAUUUUUCUUCCAUUCUU-UUUUUUUUUGCUUGU-GCAGUUUAACGCAUAAUUUAUUAUGCGAGGCAAAAAUCUGCUGCUGCA .........--.......................-..............(-(((((....(((((((....))))))).((((......)))))))))) ( -17.80) >DroSec_CAF1 88742 92 - 1 CAUUUUCUUC-CCCUAUAAUUUAAUUUCAGUU---U-U-UUUUCGCCUUCCCAACUGCAACGCAUAAUUUAUUAUGCGGCGCA-AAAUCUUGUGCACCA ..........-................(((((---.-.-.............)))))...(((((((....)))))))(((((-(....)))))).... ( -15.56) >DroEre_CAF1 101930 93 - 1 CAUUUUCUUC--CCUAUAAUUUAAUUUCUCUAU--C-UUUUUUCGCCUUCCCAGCUGCAACGCGUAAUUUAUUAUGCGGUGCA-AAGUCUUGUGCACCA ..........--.....................--.-........................((((((....))))))((((((-........)))))). ( -14.90) >DroYak_CAF1 92267 95 - 1 CAUUUUCUUC-CCCUAUAAUUUAAUUUCAGUUUU-U-UUUUUUCGCCCUCCCAACUGCAACGCAUAAUUUAUUAUGCGGAGCG-AAAUCUUGUGCACCA ..........-..................((...-.-...((((((((...(....)....((((((....)))))))).)))-)))......)).... ( -13.22) >DroAna_CAF1 116512 98 - 1 CAUUUUCUUUUUUCUAUAAUUUAAUUGAAGUUCCUU-UUUUUUGGGGCACCCCACUGCGUCACAUAAUUUAUUAUUCGAGGCAGAAAUCUGCUUCACCA .........................((((((.....-.....((((....))))((((.((..((((....))))..)).))))......))))))... ( -19.80) >consensus CAUUUUCUUC_CCCUAUAAUUUAAUUUCAGUUU__U_UUUUUUCGCCUUCCCAACUGCAACGCAUAAUUUAUUAUGCGGAGCA_AAAUCUUGUGCACCA ............................................((.........(((..(((((((....)))))))..)))..........)).... ( -9.57 = -9.38 + -0.19)

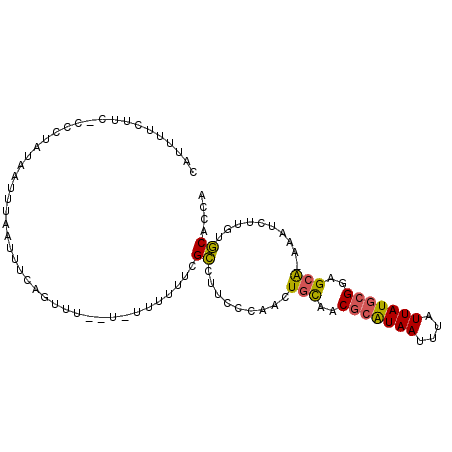
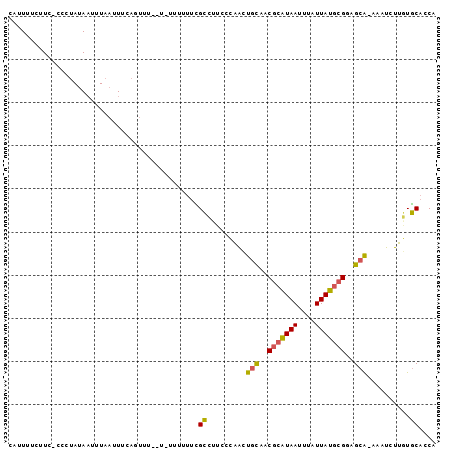
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:30 2006