| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,857,223 – 12,857,320 |
| Length | 97 |
| Max. P | 0.973677 |

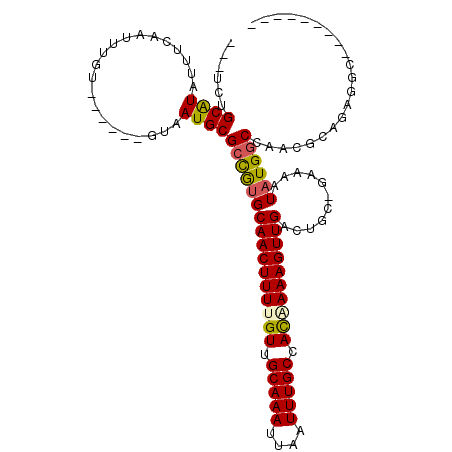
| Location | 12,857,223 – 12,857,320 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12857223 97 + 22407834 --------GGCCUCUGCGUUGGCCAUAUUUUC-GCAGUCAACUUUUGUGGCAAAUUAAUUUGCAACAAAAGUUGCACGGCGCAUUAC------ACAAAUUGAAAUAUGCAGA--- --------((((........))))........-((.((((((((((((.(((((....))))).)))))))))).)).))((((...------.(.....)....))))...--- ( -31.60) >DroPse_CAF1 46990 103 + 1 GAU---UCUGCCACUGCGUCAGCCAUAUUUUCCAGAGUCAACUUUGGUGGCAAAUUAAUUUGCAACAAAAGUUGCACAGCGCAUUAC------ACAAAUUGAAAUAUGCAGA--- ...---(((((...(((((..((((......((((((....)))))))))).........((((((....))))))..)))))...(------(.....))......)))))--- ( -28.40) >DroWil_CAF1 54367 100 + 1 --------GGGAUAUGCCGCUGCCAUAUUUUG-CCAGUCAACUUUCAUGGCAAAUUAAUUUGCAACAAAAGUUGCAUGGCGCAUUAC------AUAAAUUGAAAUACGCAAAAAU --------......(((.((.(((((..((((-(((...........))))))).......(((((....))))))))))))....(------(.....))......)))..... ( -25.30) >DroYak_CAF1 39248 95 + 1 GCGCUG----------CCUUGGCCAUAUUUUC-GCAGUCAACUUUUGUGGCAAAUUAAUUUGCAACAAAAGUUGCACGGCGCAUUAC------ACAAAUUGAAAUAUGCAGA--- ((((((----------(....)).........-...((((((((((((.(((((....))))).)))))))))).))))))).....------...................--- ( -28.30) >DroAna_CAF1 61843 100 + 1 -----------AUUUGGCUUGGUCAUAUUUUU-GCAGUCAACUUUCGUGGCAAAUUAAUUUGCAACAAAAGUUGCGCAUCGCAUUAUACUCGUACAAAUUGAAAUAUGCAGA--- -----------...((((...))))......(-((.(.(((((((.((.(((((....))))).)).)))))))))))..((((.....(((.......)))...))))...--- ( -19.40) >DroPer_CAF1 63768 103 + 1 GAU---UCUGCCACUGCGUCAGCCAUAUUUUCCAGAGUCAACUUUGGUGGCAAAUUAAUUUGCAACAAAAGUUGCACAGCGCAUUAC------ACAAAUUGAAAUAUGCAGA--- ...---(((((...(((((..((((......((((((....)))))))))).........((((((....))))))..)))))...(------(.....))......)))))--- ( -28.40) >consensus _________GCCUCUGCCUCGGCCAUAUUUUC_GCAGUCAACUUUCGUGGCAAAUUAAUUUGCAACAAAAGUUGCACAGCGCAUUAC______ACAAAUUGAAAUAUGCAGA___ .....................(.((((((((........((((((.((.(((((....))))).)).))))))((.....))..................)))))))))...... (-14.62 = -14.95 + 0.33)

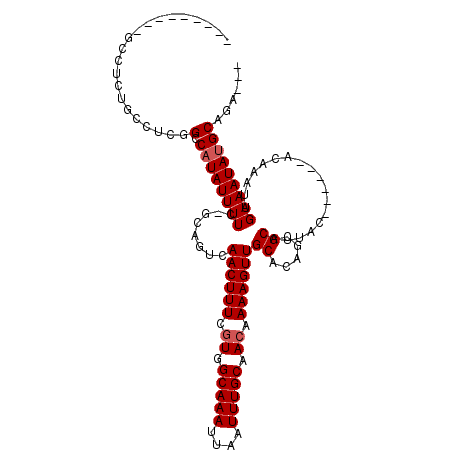
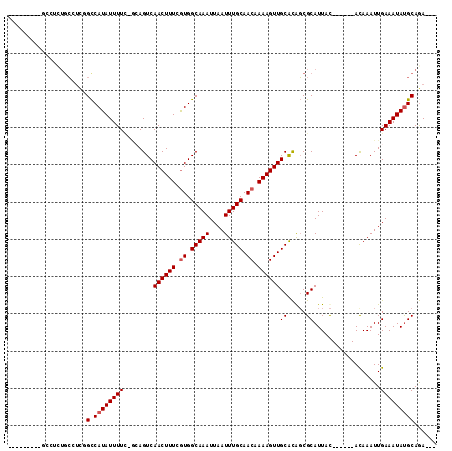
| Location | 12,857,223 – 12,857,320 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -18.91 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12857223 97 - 22407834 ---UCUGCAUAUUUCAAUUUGU------GUAAUGCGCCGUGCAACUUUUGUUGCAAAUUAAUUUGCCACAAAAGUUGACUGC-GAAAAUAUGGCCAACGCAGAGGCC-------- ---..((((((........)))------)))...(((.((.((((((((((.(((((....))))).)))))))))))).))-).......((((........))))-------- ( -32.50) >DroPse_CAF1 46990 103 - 1 ---UCUGCAUAUUUCAAUUUGU------GUAAUGCGCUGUGCAACUUUUGUUGCAAAUUAAUUUGCCACCAAAGUUGACUCUGGAAAAUAUGGCUGACGCAGUGGCAGA---AUC ---(((((((((........))------))....(((((((((((....)))))..........(((((((.((....)).)))......))))....)))))))))))---... ( -28.80) >DroWil_CAF1 54367 100 - 1 AUUUUUGCGUAUUUCAAUUUAU------GUAAUGCGCCAUGCAACUUUUGUUGCAAAUUAAUUUGCCAUGAAAGUUGACUGG-CAAAAUAUGGCAGCGGCAUAUCCC-------- ....(((((((((.((.....)------).)))))((((..(((((((..(.(((((....))))).)..)))))))..)))-)........))))...........-------- ( -28.50) >DroYak_CAF1 39248 95 - 1 ---UCUGCAUAUUUCAAUUUGU------GUAAUGCGCCGUGCAACUUUUGUUGCAAAUUAAUUUGCCACAAAAGUUGACUGC-GAAAAUAUGGCCAAGG----------CAGCGC ---..((((((........)))------)))..(((((((.((((((((((.(((((....))))).))))))))))...))-)........((....)----------).)))) ( -31.10) >DroAna_CAF1 61843 100 - 1 ---UCUGCAUAUUUCAAUUUGUACGAGUAUAAUGCGAUGCGCAACUUUUGUUGCAAAUUAAUUUGCCACGAAAGUUGACUGC-AAAAAUAUGACCAAGCCAAAU----------- ---....(((((((....(((((....))))).....(((.((((((((((.(((((....))))).))))))))))...))-).)))))))............----------- ( -24.20) >DroPer_CAF1 63768 103 - 1 ---UCUGCAUAUUUCAAUUUGU------GUAAUGCGCUGUGCAACUUUUGUUGCAAAUUAAUUUGCCACCAAAGUUGACUCUGGAAAAUAUGGCUGACGCAGUGGCAGA---AUC ---(((((((((........))------))....(((((((((((....)))))..........(((((((.((....)).)))......))))....)))))))))))---... ( -28.80) >consensus ___UCUGCAUAUUUCAAUUUGU______GUAAUGCGCCGUGCAACUUUUGUUGCAAAUUAAUUUGCCACAAAAGUUGACUGC_GAAAAUAUGGCCAACGCAGAGGC_________ ......((((.....................))))((((((((((((((((.(((((....))))).))))))))))...........))))))..................... (-18.91 = -18.80 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:01 2006