
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,856,552 – 12,856,676 |
| Length | 124 |
| Max. P | 0.990438 |

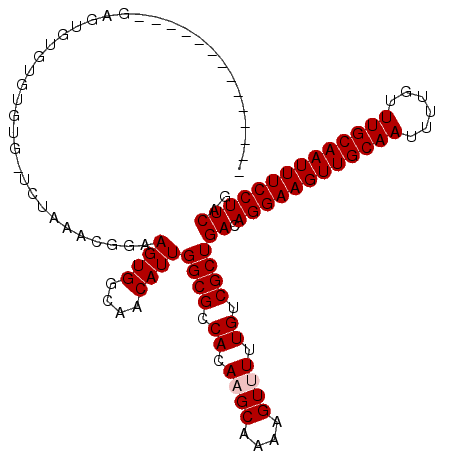
| Location | 12,856,552 – 12,856,652 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12856552 100 + 22407834 --------------GAGUGUAUGUGUG-UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAG --------------...(((.((((((-((...((....))(....)...)))).)))).))).............((((.((((((((((((......)))))))))))))))) ( -33.50) >DroVir_CAF1 40329 100 + 1 --------------GAGUGUGUGUGUGCUCUAAACGGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAG --------------.((((((((.(((((....((....))(....)...)))))))))-(((((....)))))))))((.((((((((((((......)))))))))))))).. ( -35.00) >DroPse_CAF1 46549 96 + 1 --G----------------UGUGUGUG-UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAG --.----------------((((.(((-((...((....))(....)...))))))))).(((((....)))))..((((.((((((((((((......)))))))))))))))) ( -34.90) >DroSec_CAF1 37086 100 + 1 --------------GAGUGUAUGUGUG-UCUAAGCGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAG --------------...(((.((((((-((...((....))(....)...)))).)))).))).............((((.((((((((((((......)))))))))))))))) ( -33.80) >DroMoj_CAF1 71820 100 + 1 --------------GUGUGUGUAUGUUCUCUAAACGGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAC --------------(((.(((((((((((....((....)))).)))))..))))))).-(((((....)))))...(((.((((((((((((......))))))))))))))). ( -30.80) >DroAna_CAF1 61174 114 + 1 UUGUGUGUGCUUAGAUGUGUGUUGGUG-UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCGG .......(((((.(.((.((((..(((-(....((....))....))))..)))))))))))).............((((.((((((((((((......)))))))))))))))) ( -40.50) >consensus ______________GAGUGUGUGUGUG_UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAG ......................................((((....))))((((.((.((((....)))).)).))))((.((((((((((((......)))))))))))))).. (-28.27 = -28.60 + 0.33)


| Location | 12,856,552 – 12,856,652 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12856552 100 - 22407834 CUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA-CACACAUACACUC-------------- ....(((((.((((((......)))))).)))))((((((((.........))))).((((((.......)))))).........))-)............-------------- ( -25.20) >DroVir_CAF1 40329 100 - 1 CUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCCGUUUAGAGCACACACACACUC-------------- (((((((((.((((((......)))))).))))).)))).............(((-(((((((.......))))))...(.....)))))...........-------------- ( -25.10) >DroPse_CAF1 46549 96 - 1 CUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA-CACACACA----------------C-- ....(((((.((((((......)))))).)))))((((((((.........))))).((((((.......)))))).........))-).......----------------.-- ( -25.20) >DroSec_CAF1 37086 100 - 1 CUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGCUUAGA-CACACAUACACUC-------------- ....(((((.((((((......)))))).)))))(((((((.((((....))))...((((((.......))))))...))))..))-)............-------------- ( -26.90) >DroMoj_CAF1 71820 100 - 1 GUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCCGUUUAGAGAACAUACACACAC-------------- (((.(((((.((((((......)))))).))))).(((((((.........))))-)((((((.......)))))).........)).......)))....-------------- ( -25.40) >DroAna_CAF1 61174 114 - 1 CCGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA-CACCAACACACAUCUAAGCACACACAA ..(((((((.((((((......)))))).))))).))(((((.........))))).((((((.......))))))....(((((((-............)))))))........ ( -27.80) >consensus CUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA_CACACACACACUC______________ ..(((((((.((((((......)))))).))))).))((((.((((....))))...((((((.......))))))...))))................................ (-22.69 = -22.55 + -0.14)

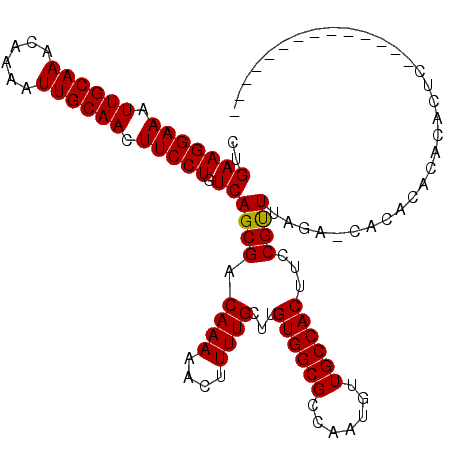

| Location | 12,856,572 – 12,856,676 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -33.01 |
| Energy contribution | -32.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12856572 104 + 22407834 GGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUG------CUCAAAGGGU-------- ...((((....))))((((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.))))------))........-------- ( -42.10) >DroVir_CAF1 40350 99 + 1 GGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGUG-CACAUUU-----G------CCAAAAGUGU-------- ....(((((.........))))).-(((((......(((((((((.((((((((((((......))))))))))))))))))-.))))))-----)------).........-------- ( -37.20) >DroGri_CAF1 58967 107 + 1 GGAAGUGGCAACGUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGUG-CACAUUU-----G------CCAAAACUGGGUAGGGGG ((...((((((.((((......))-)).........(((((((((.((((((((((((......))))))))))))))))))-.))).))-----)------)))...)).......... ( -38.90) >DroWil_CAF1 54016 112 + 1 GGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCGACACAUUUUCGGUGAGGAGUUUCAAAGGGA-------- ....(((((.........))))).............(((((((((.((((((((((((......)))))))))))))))))))))...(..(..((((....))))..)..)-------- ( -43.10) >DroMoj_CAF1 71841 99 + 1 GGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCACUG-CACAUUU-----G------CCAAAAGUUG-------- ((((((((((.....((((.((.(-((....)))..)).))))((.((((((((((((......))))))))))))))..))-).)))))-----.------))........-------- ( -31.70) >DroAna_CAF1 61208 103 + 1 GGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCGGCG-CACAUUU-UGGAG------ACCAAG-AAG-------- ....(((((.........))))).............(((((((((.((((((((((((......))))))))))))))))))-.)))(((-(((..------.)))))-)..-------- ( -38.90) >consensus GGAAGUGGCAACAUUGGCGCCACA_GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG_CACAUUU_____G______CCAAAAGUGG________ ....(((((.........))))).............((.((((((.((((((((((((......)))))))))))))))))).))................................... (-33.01 = -32.78 + -0.22)


| Location | 12,856,572 – 12,856,676 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12856572 104 - 22407834 --------ACCCUUUGAG------CACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCC --------.......(((------((...-.....((-(((((((((((.((((((......)))))).))))).)))))).))........)))))((((((.......)))))).... ( -36.39) >DroVir_CAF1 40350 99 - 1 --------ACACUUUUGG------C-----AAAUGUG-CACUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCC --------........((------(-----(((.((.-(.(((((((((.((((((......)))))).))))).)))).)......)).)))))-)((((((.......)))))).... ( -31.00) >DroGri_CAF1 58967 107 - 1 CCCCCUACCCAGUUUUGG------C-----AAAUGUG-CACUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAACGUUGCCACUUCC ................((------(-----(((.((.-(.(((((((((.((((((......)))))).))))).)))).)......)).)))))-)((((((.......)))))).... ( -31.00) >DroWil_CAF1 54016 112 - 1 --------UCCCUUUGAAACUCCUCACCGAAAAUGUGUCGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCC --------...................(((((.(.((((((((((((((.((((((......)))))).))))).)))))))))...).)))))...((((((.......)))))).... ( -37.00) >DroMoj_CAF1 71841 99 - 1 --------CAACUUUUGG------C-----AAAUGUG-CAGUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCC --------.......(((------(-----((..(((-(.....(((((.((((((......)))))).)))))..((((((.........))))-))..)))).....))))))..... ( -29.70) >DroAna_CAF1 61208 103 - 1 --------CUU-CUUGGU------CUCCA-AAAUGUG-CGCCGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCC --------...-..((((------...((-...((.(-(((((((((((.((((((......)))))).))))).)).((((((((....)))).))))))))))).))..))))..... ( -29.80) >consensus ________CCACUUUGGG______C_____AAAUGUG_CACUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC_UGUGGCGCCAAUGUUGCCACUUCC ...................................((.(.(((((((((.((((((......)))))).))))).)))).).)).............((((((.......)))))).... (-22.88 = -23.22 + 0.33)
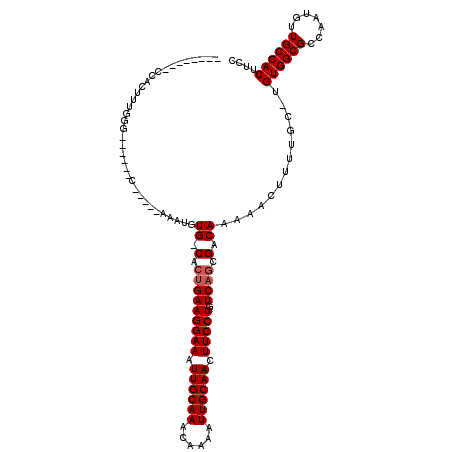
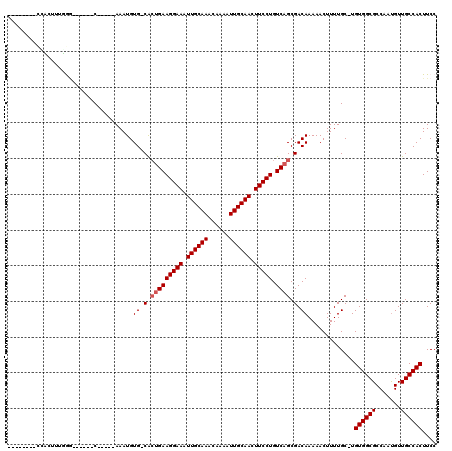
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:59 2006