| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,851,532 – 12,851,629 |
| Length | 97 |
| Max. P | 0.878429 |

| Location | 12,851,532 – 12,851,629 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.29 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
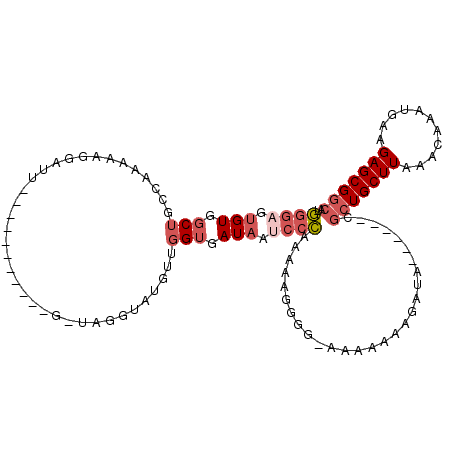
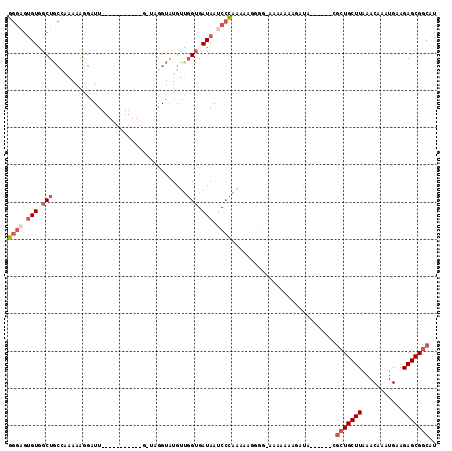
>2L_DroMel_CAF1 12851532 97 - 22407834 GGGAGUGUGGCUGCCAAAAAGGAUU-----------CGUAGGUAUGUUGGUGAUAAUCCCAAAAAGGGG-GAAAAAAGAUA------CGCUGCUUAAACAAAUGAAGAGCGGCAU ((.((.....)).))..........-----------.....((((.((..(.....((((.....))))-..)..)).)))------)(((((((...........))))))).. ( -22.50) >DroSec_CAF1 32082 97 - 1 GGGAGUGUGGCUGCCAAAAAGGAUU-----------GGUAGGUAUGUUGGUGAUAAUCCCAAAAAGGGG-AAAAAAAGAUA------CGCUGCUUAAACAAAUGAAGAGCGGCAU ...((..((.(((((((......))-----------))))).))..))........((((......)))-)..........------.(((((((...........))))))).. ( -27.30) >DroSim_CAF1 34607 97 - 1 GGGAGUGUGGCUGCCAAAAAGGAUU-----------GCUAGGUAUGUUGGUGAUAAUCCCAAAAAGGGG-GAAAAAAGAUA------CGCUGCUUAAACAAAUGAAGAGCGGCAU .......((((..((.....))...-----------)))).((((.((..(.....((((.....))))-..)..)).)))------)(((((((...........))))))).. ( -23.50) >DroEre_CAF1 38023 90 - 1 GGGAGUGUGGCUGCC-AAAAGGAUU---------------GGUGUGUUGGGGAUAAUCGCAAAGA---GCAAAAAAAGAUA------CGCUGCUUAAACAAAUGAAGAGCGGCAU ....((((....(((-((.....))---------------)))((.(((.(......).)))...---))........)))------)(((((((...........))))))).. ( -18.80) >DroYak_CAF1 33372 89 - 1 GG----GUGGCUGCA-AAAAGGAUU---------------GGUAUGUCGGUGAUAAUCCCGAAAAGGGGAAAAAAAAUAAA------CGCUGCUUAAACAAAUGAAGAGCGGCAU ..----((.((((((-.........---------------....)).)))).))..((((......))))...........------.(((((((...........))))))).. ( -19.02) >DroAna_CAF1 56199 112 - 1 UGGUGUGUGGCUUAGA-AAGGGGUUCCAUGAGGUUUGAGUGGCGUGGU-GUGAUAAUCCAACGAAAAAA-GAAAAAAUUUAUAAAAGUAAUGCUUAAACAAAUGAAGAGCGACAU ....((((.((((.((-(.....)))(((...((((((((..(((((.-........)).)))......-........((((....)))).))))))))..)))..)))).)))) ( -20.00) >consensus GGGAGUGUGGCUGCCAAAAAGGAUU___________G_UAGGUAUGUUGGUGAUAAUCCCAAAAAGGGG_AAAAAAAGAUA______CGCUGCUUAAACAAAUGAAGAGCGGCAU ((((.(((.(((....................................))).))).))))............................(((((((...........))))))).. (-12.06 = -13.29 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:56 2006