
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,847,249 – 12,847,403 |
| Length | 154 |
| Max. P | 0.706894 |

| Location | 12,847,249 – 12,847,340 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.92 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
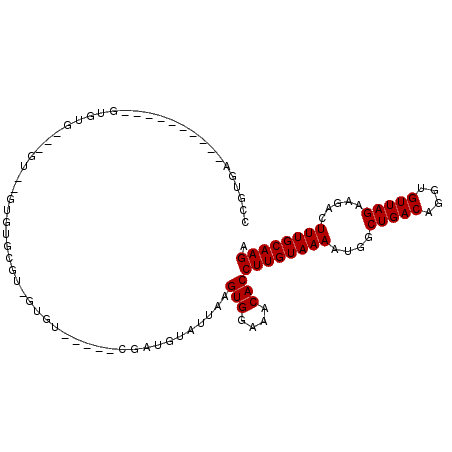
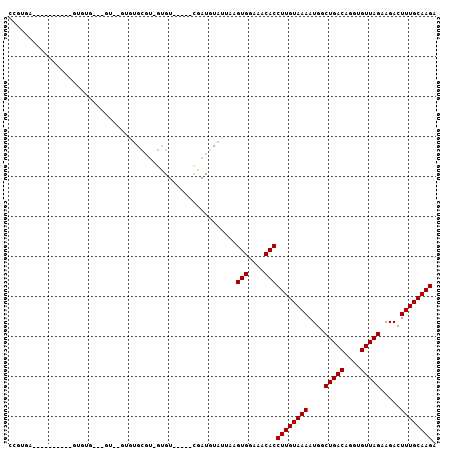
>2L_DroMel_CAF1 12847249 91 + 22407834 CCGUGA----------GUGUGUUCGUAUGUGUGCGU-GUGU-----UGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA .(....----------).(((((..(((..((((((-....-----..))))))...)))..)))))((((((((....(((((....))))).....)))))))). ( -22.70) >DroGri_CAF1 45830 90 + 1 CUGUCUCUUUCUAGCC------------GUGUGUGUGAUGU-----CGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA ..((((...(((((((------------.(((..((.((..-----.....(((...(((....)))...)))..)).))..))).).))))))))))......... ( -20.60) >DroEre_CAF1 33661 91 + 1 CCGUGA----------GUGUGUGUGUGAGUGUGCGU-GUGU-----CGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA .((...----------(..((..(......)..)).-.)..-----)).........(((....)))((((((((....(((((....))))).....)))))))). ( -22.10) >DroYak_CAF1 28870 85 + 1 CCGUGA----------GUGUAUGUGU------GUGU-GUGU-----CGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA .((...----------(..(((....------))).-.)..-----)).........(((....)))((((((((....(((((....))))).....)))))))). ( -19.40) >DroAna_CAF1 52147 92 + 1 CACUAG----------GUGUG----GGAGUGUGCUG-GUGUUGGUGCGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA ((((((----------(..(.----...)..).)))-)))(((((((...)))))))(((....)))((((((((....(((((....))))).....)))))))). ( -24.40) >DroPer_CAF1 48872 73 + 1 ------------------------GG----CUGUGU-GCGU-----CGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA ------------------------.(----(.....-))..-----...........(((....)))((((((((....(((((....))))).....)))))))). ( -18.70) >consensus CCGUGA__________GUGUG___GU__GUGUGCGU_GUGU_____CGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGA .........................................................(((....)))((((((((....(((((....))))).....)))))))). (-17.20 = -17.20 + -0.00)

| Location | 12,847,300 – 12,847,403 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
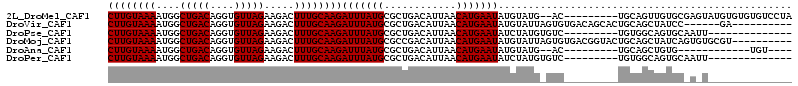
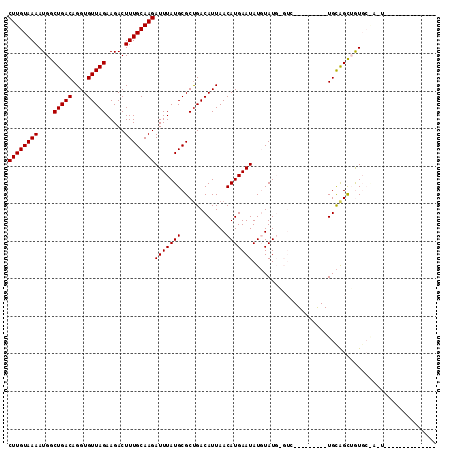
>2L_DroMel_CAF1 12847300 103 + 22407834 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUG--AC---------UGCAGUUGUGCGAGUAUGUGUGUGUCCUA ((((((((....(((((....))))).....))))))))...........(((((..(((((....((((..--((---------....))..))))..)))))..)))))... ( -26.80) >DroVir_CAF1 28247 98 + 1 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUUAGUGUGACAGCACUGCAGCUAUCC------GA---------- ........(((((((.(((...............(((.......)))((((((((((((((....))))..))))))..)))).))))))))))..------..---------- ( -24.40) >DroPse_CAF1 33352 91 + 1 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUCUAUGUGUC---------UGUGGCAGUGCAAUU-------------- ((((((((....(((((....))))).....)))))))).....(((((((.(((..(((((......)))))...---------.))).)))))))...-------------- ( -28.00) >DroMoj_CAF1 59433 104 + 1 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCCGACAUUAACAUGAAUAUGUAUUAGUGUGACGGUACUGCAGCUAUCAGUGUGCGU---------- ((((((((....(((((....))))).....))))))))....(((((((.((((((((((....))))..))))))((..((......))..)).).))))))---------- ( -25.40) >DroAna_CAF1 52199 87 + 1 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUG--AC---------UGCAGCUGUG------------UGU---- ((((((((....(((((....))))).....))))))))...(((((((((.(((((((((....)))).))--).---------)))))).)))------------)).---- ( -21.30) >DroPer_CAF1 48905 91 + 1 CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUCUAUGUGUC---------UGUGGCAGUGCAAUU-------------- ((((((((....(((((....))))).....)))))))).....(((((((.(((..(((((......)))))...---------.))).)))))))...-------------- ( -28.00) >consensus CUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUG_GUC_________UGCAGCUGUGC_A_U______________ ((((((((....(((((....))))).....))))))))(((((((............)))))))................................................. (-14.30 = -14.30 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:55 2006