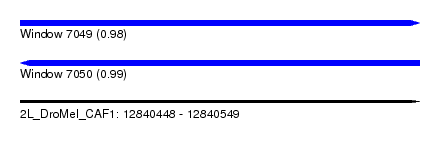
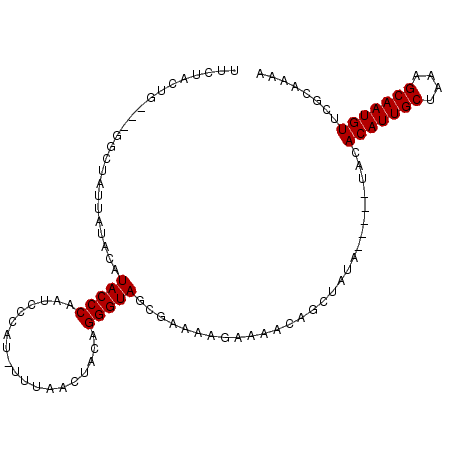
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,840,448 – 12,840,549 |
| Length | 101 |
| Max. P | 0.991419 |

| Location | 12,840,448 – 12,840,549 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.76 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12840448 101 + 22407834 UUUUGCGAACAUUGCUUUAGCAAUGUGUA-----UAUAGCUGUUUUCUUUUAGCUACCCUGUAGUUAAAAAUGGGAUUGGGUAUGUAUAAUAGCCGAGCAGUAGAA ..((((..(((((((....)))))))(((-----((((.((..(..(((((((((((...))))))))))..)..)..)).))))))).........))))..... ( -28.40) >DroSec_CAF1 21238 101 + 1 UUUUGCGAACAUUGCUUUAGCAAUGUGUA-----UAUAGCUGUUUUCUUUUAGCUACCCUGUAGUUAAAUAUGGGAUUGGGUAUGUAUAAUAGCCCAUCAGUAGAA .(((((..(((((((....)))))))(((-----((((.((..(..(.(((((((((...)))))))))...)..)..)).)))))))............))))). ( -28.30) >DroSim_CAF1 23483 101 + 1 UUUUGCGAACAUUGCUUUAGCAAUGUGUA-----UAUAGCUGUUUUCUUUUCGCUACCCUGUAGUUAAAUAUGGGAUUGGGUAUGUAUAAAAGCCUGUCAGUAGAA .(((((..(((((((....)))))))(((-----((((.((..(..(.(((.(((((...))))).)))...)..)..)).)))))))............))))). ( -24.70) >DroEre_CAF1 26960 86 + 1 UCUUGCGAACAUUGCUUUAGCAAUGUGUACUCCACAUAGCUCUUUUCUUUUCAGUACCCUGUAGCGGGA-AUGGCAAUGGGUA----GU---------------AG ........(((((((....))))))).((((((.(((.(((.((..(.((.(((....))).)).)..)-).))).))))).)----))---------------). ( -24.20) >DroYak_CAF1 21991 87 + 1 UUUUGCGAACAUUGCUUUCGCAAUGUGUA-----UAUAGCUCUUUUCUUUUCAGUACCCUGUAGCUGUA-AUGACAUAGGGUA----UAAUA---------UACAA ........(((((((....)))))))(((-----(((((.......)).....(((((((((..(....-..)..))))))))----).)))---------))).. ( -21.60) >consensus UUUUGCGAACAUUGCUUUAGCAAUGUGUA_____UAUAGCUGUUUUCUUUUCGCUACCCUGUAGUUAAA_AUGGGAUUGGGUAUGUAUAAUAGCC___CAGUAGAA ........(((((((....)))))))........((((.((..((((.(((((((((...)))))))))...))))..)).))))..................... (-13.84 = -15.76 + 1.92)

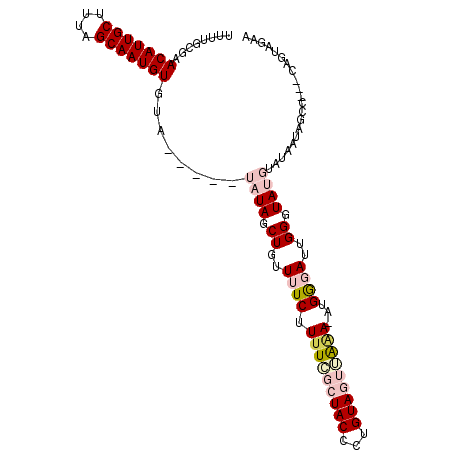
| Location | 12,840,448 – 12,840,549 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -19.89 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.31 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12840448 101 - 22407834 UUCUACUGCUCGGCUAUUAUACAUACCCAAUCCCAUUUUUAACUACAGGGUAGCUAAAAGAAAACAGCUAUA-----UACACAUUGCUAAAGCAAUGUUCGCAAAA ......(((..((((........(((((...................))))).............))))...-----...(((((((....)))))))..)))... ( -17.01) >DroSec_CAF1 21238 101 - 1 UUCUACUGAUGGGCUAUUAUACAUACCCAAUCCCAUAUUUAACUACAGGGUAGCUAAAAGAAAACAGCUAUA-----UACACAUUGCUAAAGCAAUGUUCGCAAAA .........((((.(((.....)))))))..(((.............)))(((((..........)))))..-----...(((((((....)))))))........ ( -17.42) >DroSim_CAF1 23483 101 - 1 UUCUACUGACAGGCUUUUAUACAUACCCAAUCCCAUAUUUAACUACAGGGUAGCGAAAAGAAAACAGCUAUA-----UACACAUUGCUAAAGCAAUGUUCGCAAAA .....(((.....(((((.....(((((...................)))))...)))))....))).....-----...(((((((....)))))))........ ( -17.81) >DroEre_CAF1 26960 86 - 1 CU---------------AC----UACCCAUUGCCAU-UCCCGCUACAGGGUACUGAAAAGAAAAGAGCUAUGUGGAGUACACAUUGCUAAAGCAAUGUUCGCAAGA .(---------------((----(.(((((.(((..-.(((......)))..((....))....).)).))).)))))).(((((((....)))))))((....)) ( -24.00) >DroYak_CAF1 21991 87 - 1 UUGUA---------UAUUA----UACCCUAUGUCAU-UACAGCUACAGGGUACUGAAAAGAAAAGAGCUAUA-----UACACAUUGCGAAAGCAAUGUUCGCAAAA ..(((---------(((..----((((((..((...-....))...))))))((....)).........)))-----)))(((((((....)))))))........ ( -23.20) >consensus UUCUACUG___GGCUAUUAUACAUACCCAAUCCCAU_UUUAACUACAGGGUAGCGAAAAGAAAACAGCUAUA_____UACACAUUGCUAAAGCAAUGUUCGCAAAA .......................(((((...................)))))............................(((((((....)))))))........ (-13.31 = -13.31 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:52 2006