
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,832,481 – 12,832,582 |
| Length | 101 |
| Max. P | 0.803654 |

| Location | 12,832,481 – 12,832,582 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
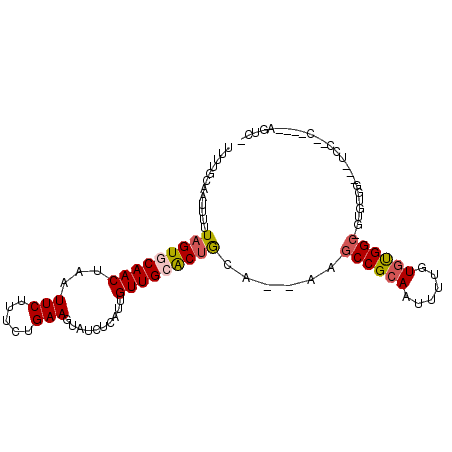
>2L_DroMel_CAF1 12832481 101 - 22407834 UUUUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGGACUGCGA-AAGCCGCAAUUUAGUGCGGC-GUAUGGGGUUCCGCCUGCCACUGU ............(((((((.....(((.....)))..........((.(((((.((.-..((((((......))))))-...)).))))).)).)).))))). ( -25.10) >DroVir_CAF1 13553 93 - 1 UUUUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGCACUGCA--AAGCCGCAAUUUUGUGUGGC-GUGUGGAUAUCC--C----AGUU- .((..((.(((.(((((((((...(((.....)))..........))))))))).)--))((((((......))))))-.))..)).....--.----....- ( -24.72) >DroGri_CAF1 31241 93 - 1 UUUUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGCACUGCA--AAGCCGCAAUUCUGUGUGGC-GUGUGGAUAUCG--C----AGUC- ..((((......(((((((((...(((.....)))..........)))))))))((--..((((((......))))))-...))......)--)----))..- ( -25.42) >DroWil_CAF1 18117 98 - 1 UUUUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGCACUGCAA-AAGCCGCAAUUUUGUAUGGCCGUCUG----UCCUCCGGCCAGUUG ..((((..(((((((((((((...(((.....)))..........))))))))).))-))...)))).......((((((....----.....)))))).... ( -25.72) >DroMoj_CAF1 38621 84 - 1 UUUAGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGCACUGCA--AAGCCGCAAUUUCGUGUGGC-AUGCA-----CC--C--------- ....(((.(((.(((((((((...(((.....)))..........))))))))).)--))((((((......))))))-.))).-----..--.--------- ( -24.62) >DroAna_CAF1 38954 99 - 1 UUCUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUUAUUGUUGGGCCACAAAAAACCGCAAUUUAGUGCGGC-GUAUGCGGGC---UAUGCCACUCU .((((((.(((((.(((...(((((.......((....)).....)))))..))).)))))(((((......))))).-...)))))).---........... ( -19.10) >consensus UUUUGCAAUUUUUAGUGCAACUAAUUCUUUCUGAAGUAUCUCAUUGUUGCACUGCA__AAGCCGCAAUUUUGUGUGGC_GUGUGG___UCC__C____AGUC_ ............(((((((((...(((.....)))..........)))))))))......((((((......))))))......................... (-15.70 = -16.04 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:47 2006