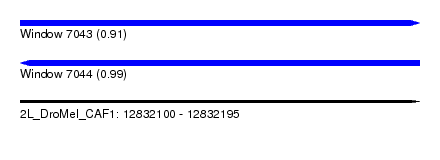
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,832,100 – 12,832,195 |
| Length | 95 |
| Max. P | 0.991151 |

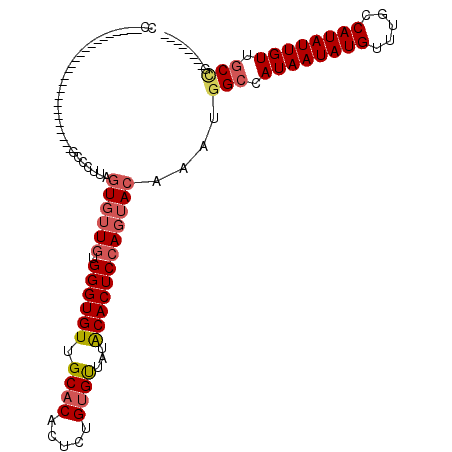
| Location | 12,832,100 – 12,832,195 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.20 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -12.84 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
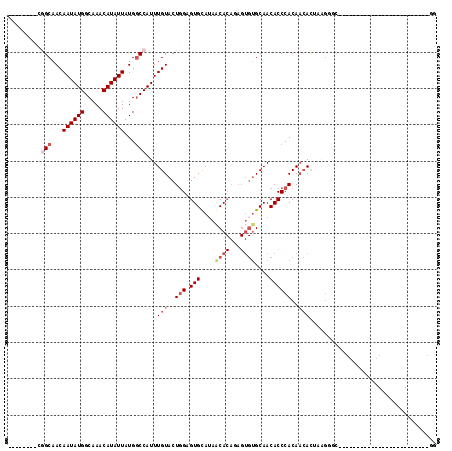
>2L_DroMel_CAF1 12832100 95 + 22407834 --------CGGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUGGAGUGUAUAACACAGAGUGUGCAACACCCACAACACUAAGGGCCACCAAA---------------UGAGG --------.((....((((((.....)))))).(((((...(((..(((.((((...((((...))))...)))))))..))).....)))))))...---------------..... ( -26.00) >DroSec_CAF1 12921 110 + 1 --------CGGCAACAAUAUGGCAAACAUAUUAUGUCCAUUUGUACUGGAGUGUAUAACACAGAGUGUGCAACACCCACAACACUAAGGGUCACCAGAAGGGGCAUGGGCACUUGGGG --------.(....)((((((.....)))))).(((((((.(((..(((.((((...((((...))))...)))))))..)))......(((.((....))))))))))))....... ( -29.00) >DroSim_CAF1 13586 109 + 1 --------CGGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUGGAGUGUAUAACACAAAGUGUGCAACACCCACAACACUAAGGGCCACCAGGAG-GGCAUGGGCACUUGGGG --------.(((...((((((.....))))))...)))...(((..(((.((((...((((...))))...)))))))..)))(((((.(((.((....)-).....))).))))).. ( -33.30) >DroEre_CAF1 18665 78 + 1 --------CGGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUGGAGUGCAGAGCACAGAGUGUGCAACACCCACAACAC--------------------------------AG --------.(((...((((((.....))))))...))).(((((((....)))))))((((.....))))..............--------------------------------.. ( -22.60) >DroYak_CAF1 13307 92 + 1 UAGAGUGCAAGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUGGAGUGCAUAACACAGAGUGUGCAACACCCACAACAUCGGGG-U-------------------------GG ...(((((((((...((((((.....))))))...))...)))))))...(..(((........)))..)..(((((.(......))))-)-------------------------). ( -26.30) >DroAna_CAF1 38541 76 + 1 --------CUGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUCGAGUGCAUAACA-------GGCAACACCCACAUAACACAGAGC--------------------------- --------(((....((((((.....))))))...(((...(((((....))))).....-------)))...............)))...--------------------------- ( -14.40) >consensus ________CGGCAACAAUAUGGCAAACAUAUUAUGGCCAUUUGUACUGGAGUGCAUAACACAGAGUGUGCAACACCCACAACACUAAGGGC_________________________GG .........(((...((((((.....))))))...)))...(((..(((.(((....((((...))))....))))))..)))................................... (-12.84 = -14.27 + 1.43)

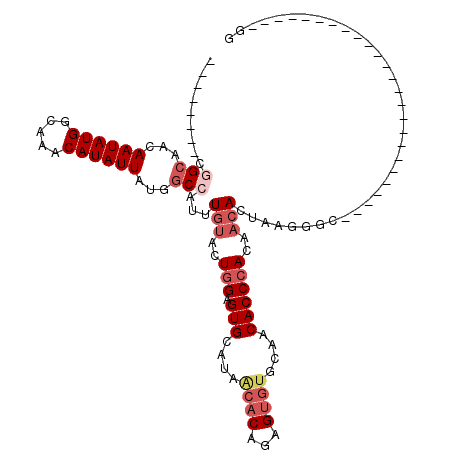
| Location | 12,832,100 – 12,832,195 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.20 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12832100 95 - 22407834 CCUCA---------------UUUGGUGGCCCUUAGUGUUGUGGGUGUUGCACACUCUGUGUUAUACACUCCAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCCG-------- .....---------------...(((((((.((.((((((.((((((.((((.....))))...)))))))))))).)).))))((((((((.....)))))))).))).-------- ( -32.40) >DroSec_CAF1 12921 110 - 1 CCCCAAGUGCCCAUGCCCCUUCUGGUGACCCUUAGUGUUGUGGGUGUUGCACACUCUGUGUUAUACACUCCAGUACAAAUGGACAUAAUAUGUUUGCCAUAUUGUUGCCG-------- .......................((..((.....((((((.((((((.((((.....))))...))))))))))))..((((.((.........))))))...))..)).-------- ( -27.10) >DroSim_CAF1 13586 109 - 1 CCCCAAGUGCCCAUGCC-CUCCUGGUGGCCCUUAGUGUUGUGGGUGUUGCACACUUUGUGUUAUACACUCCAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCCG-------- ....(((.(((...(((-.....)))))).))).((((((.((((((.((((.....))))...))))))))))))....(((.((((((((.....)))))))).))).-------- ( -35.00) >DroEre_CAF1 18665 78 - 1 CU--------------------------------GUGUUGUGGGUGUUGCACACUCUGUGCUCUGCACUCCAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCCG-------- .(--------------------------------((((((.((((((.((((.....))))...)))))))))))))...(((.((((((((.....)))))))).))).-------- ( -30.50) >DroYak_CAF1 13307 92 - 1 CC-------------------------A-CCCCGAUGUUGUGGGUGUUGCACACUCUGUGUUAUGCACUCCAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCUUGCACUCUA ..-------------------------.-......(((..(((((((.((((.....))))...)))).)))..)))...(((.((((((((.....)))))))).)))......... ( -23.20) >DroAna_CAF1 38541 76 - 1 ---------------------------GCUCUGUGUUAUGUGGGUGUUGCC-------UGUUAUGCACUCGAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCAG-------- ---------------------------((((.((((...(..((.....))-------..)...))))..)))).......((.((((((((.....)))))))).))..-------- ( -21.60) >consensus CC_________________________GCCCUUAGUGUUGUGGGUGUUGCACACUCUGUGUUAUACACUCCAGUACAAAUGGCCAUAAUAUGUUUGCCAUAUUGUUGCCG________ ..................................((((((.((((((.((((.....))))...))))))))))))....(((.((((((((.....)))))))).)))......... (-22.81 = -23.52 + 0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:46 2006