| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,816,037 – 12,816,190 |
| Length | 153 |
| Max. P | 0.941445 |

| Location | 12,816,037 – 12,816,152 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -49.65 |
| Consensus MFE | -45.53 |
| Energy contribution | -46.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12816037 115 + 22407834 -----GAGCUUCGGGGUGGCGGGCGGUUGAGCCCAGACAGUUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCA -----...(....).(.((((((.(((((.((((((.(((.(((.((((((.....))))))...))).)))...)))))).((((((.....)))).)).......)))))))))))). ( -50.20) >DroSec_CAF1 81991 115 + 1 -----GAGCUUCGGGGUGGCGGGCGGUUGAGCCUAGACAGUUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCA -----...(....).(.((((((.(((((.(((.(((..((....((((((.....))))))..))..))))))..(((((.((((((.....)))).)).))))).)))))))))))). ( -49.40) >DroSim_CAF1 85311 115 + 1 -----GAGCUUCGGGGUGGCGGGCGGUUGAGCCCAGACAGUUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCAGUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCA -----........((((((.((((......)))).....(((.((((.((((.(((((((.(((...))))))).)))))))((((((.....)))).)))))).))).....)))))). ( -53.00) >DroYak_CAF1 85819 119 + 1 GUCCGGAGCUUUG-GGUGGCGGGCGGUUGAGCCCAGACAGUUUGGCAUAGCCGCACGCCAUGAUAUAAUCUCGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCA ....((.((...(-(((((((.(((((((.(((..........))).))))))).)))))................(((((.((((((.....)))).)).)))))......))))))). ( -46.00) >consensus _____GAGCUUCGGGGUGGCGGGCGGUUGAGCCCAGACAGUUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCA ...............(.((((((.(((((.(((.(((..((....((((((.....))))))..))..))))))..(((((.((((((.....)))).)).))))).)))))))))))). (-45.53 = -46.03 + 0.50)


| Location | 12,816,037 – 12,816,152 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -47.10 |
| Consensus MFE | -45.45 |
| Energy contribution | -45.33 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12816037 115 - 22407834 UGGGCGGGAGUUGUUAGGCAGCGCGUAAUAUAUACGGCGGCCCAGUGCCAGAUUAUAUCAUGGCGUGCGGCCAUGCCAAACUGUCUGGGCUCAACCGCCCGCCACCCCGAAGCUC----- .(((..((((..(((.((((((.((((.....))))))((((((.((((((((...))).))))))).)))).)))).)))...))((((......)))).))..))).......----- ( -47.60) >DroSec_CAF1 81991 115 - 1 UGGGCGGGAGUUGUUAGGCAGCGCGUAAUAUAUACGGCGGCCCAGUGCCAGAUUAUAUCAUGGCGUGCGGCCAUGCCAAACUGUCUAGGCUCAACCGCCCGCCACCCCGAAGCUC----- .((((((((((..((((((((...(((.....)))(((((((((.((((((((...))).))))))).))))..)))...))))))))))))..))))))...............----- ( -47.40) >DroSim_CAF1 85311 115 - 1 UGGGCGGGAGUUGUUAGGCAGCGCGUAAUAUAUACGGCGGCCCACUGCCAGAUUAUAUCAUGGCGUGCGGCCAUGCCAAACUGUCUGGGCUCAACCGCCCGCCACCCCGAAGCUC----- .(((..((((..(((.((((((.((((.....))))))(((((((.(((((((...))).))))))).)))).)))).)))...))((((......)))).))..))).......----- ( -49.40) >DroYak_CAF1 85819 119 - 1 UGGGCGGGAGUUGUUAGGCAGCGCGUAAUAUAUACGGCGGCCCAGUGCGAGAUUAUAUCAUGGCGUGCGGCUAUGCCAAACUGUCUGGGCUCAACCGCCCGCCACC-CAAAGCUCCGGAC ..((((((.((((.......((.((((.....))))))(((((((.((((.......)).(((((((....)))))))....)))))))))))))..))))))..(-(........)).. ( -44.00) >consensus UGGGCGGGAGUUGUUAGGCAGCGCGUAAUAUAUACGGCGGCCCAGUGCCAGAUUAUAUCAUGGCGUGCGGCCAUGCCAAACUGUCUGGGCUCAACCGCCCGCCACCCCGAAGCUC_____ .((((((((((..((((((((...(((.....)))(((((((((.((((((((...))).))))))).))))..)))...))))))))))))..)))))).................... (-45.45 = -45.33 + -0.13)

| Location | 12,816,072 – 12,816,190 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12816072 118 + 22407834 UUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCACAACACCUCCUCACCGGAGGCAACUCA--UCCACAAAUGU ..(((.((((..((..((((.(((...)))))))..(((((.((((((.....)))).)).))))).........))........(((((.....)))))....)))--))))....... ( -34.60) >DroSec_CAF1 82026 118 + 1 UUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCACAACAGCUCCUCACCGGAGGCACCUCA--UCCACAAGCGU ..(((.((((((((..((((.(((...)))))))..(((((.((((((.....)))).)).)))))...................))(((.....))))))....))--))))....... ( -32.40) >DroSim_CAF1 85346 118 + 1 UUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCAGUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCACAACACCGCCUCACCAGAGGCACCUCA--UCCACAAACGU ...((((.((((.(((((((.(((...))))))).)))))))((((((.....)))).)))))).......................(((((....)))))......--........... ( -39.00) >DroYak_CAF1 85858 120 + 1 UUUGGCAUAGCCGCACGCCAUGAUAUAAUCUCGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCACAGCCCCUCCUCACCGGAGGCACCUCAAACCCCCAAAUGG (((((.......((..((...(((...)))..))..(((((.((((((.....)))).)).))))).........))........(((((.....)))))............)))))... ( -28.10) >consensus UUUGGCAUGGCCGCACGCCAUGAUAUAAUCUGGCACUGGGCCGCCGUAUAUAUUACGCGCUGCCUAACAACUCCCGCCCACAACACCUCCUCACCGGAGGCACCUCA__UCCACAAACGU ...(((..........((((.(((...)))))))..(((((.((((((.....)))).)).))))).........)))..........((((....)))).................... (-27.98 = -28.22 + 0.25)

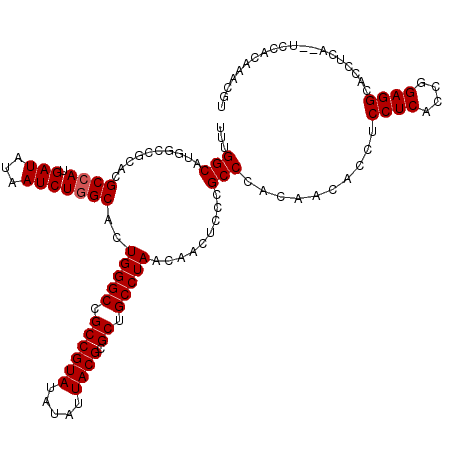
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:39 2006