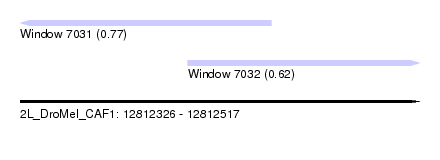
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,812,326 – 12,812,517 |
| Length | 191 |
| Max. P | 0.765701 |

| Location | 12,812,326 – 12,812,446 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12812326 120 - 22407834 AUACAAAUUCCAAUCCGAAACCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCUCUCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAACUUAAGUUC ..............(((.........((((((((.(((........))).))))))))((.(((((.(.(((.............))).).)))))))..)))................. ( -27.02) >DroSec_CAF1 78297 120 - 1 AUACGAAUUCCAAUCCGAAACCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCUCUCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAACUUAAGUUC ..............(((.........((((((((.(((........))).))))))))((.(((((.(.(((.............))).).)))))))..)))................. ( -27.02) >DroSim_CAF1 81608 120 - 1 AUACGAAUUCCAAUCCGAAACCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCUCUCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAACUUAAGUUC ..............(((.........((((((((.(((........))).))))))))((.(((((.(.(((.............))).).)))))))..)))................. ( -27.02) >DroEre_CAF1 81634 120 - 1 AUCCAAAUUCCAAUCCGAAUCCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCGCUCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAACUUAAGUUC ..............(((.........((((((((.(((........))).))))))))((.(((((.(((((.............))))).)))))))..)))................. ( -30.62) >DroYak_CAF1 82048 120 - 1 AUCCAAAUUCAAAUCCGAAGCCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCUCGCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAGCUUAAGUUC ..................((((....((((((((.(((........))).))))))))((.(((((...((((((.((...)).)))))).)))))))...))))............... ( -32.20) >consensus AUACAAAUUCCAAUCCGAAACCAAAUCGAAUCGAAUUGCUGCCAAUCAAAUCGAUUCGUGUCCUCGUCUCUCUUAUUGAUUCAUUGAGUGUCGAGGCAGCCGGCUAGUUAACUUAAGUUC ..............(((.........((((((((.(((........))).))))))))((.(((((.(.(((.............))).).)))))))..)))................. (-25.78 = -25.98 + 0.20)

| Location | 12,812,406 – 12,812,517 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12812406 111 + 22407834 AGCAAUUCGAUUCGAUUUGGUUUCGGAUUGGAAUUUGUAUUGAGGAA--------UUGAGGAAUGCUA-UCUUCGGUGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU (((((((((((((((.......)))))))))).......(..((((.--------.((((((......-))))))(((((....)))))...........))))..)...)))))..... ( -35.60) >DroSec_CAF1 78377 111 + 1 AGCAAUUCGAUUCGAUUUGGUUUCGGAUUGGAAUUCGUAUUGAGGAA--------UUGAGGAAUGCUU-UCUCCGGCGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU (((((((((((((((.......)))))))))).......(..(((((--------(((.(((......-..)))((((((....)))....)))))))..))))..)...)))))..... ( -33.10) >DroSim_CAF1 81688 112 + 1 AGCAAUUCGAUUCGAUUUGGUUUCGGAUUGGAAUUCGUAUUGAGGAA--------UUGGGGAAUGCUAUGCUCCGGCGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU (((((((((((((((.......)))))))))).......(..(((((--------(((((..(((....((....))(((....))).))).))))))..))))..)...)))))..... ( -37.00) >DroEre_CAF1 81714 119 + 1 AGCAAUUCGAUUCGAUUUGGAUUCGGAUUGGAAUUUGGAUUGAGGAAUUGAGGAAUUGAGGAAUGCCA-UCUCAGGUGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU ((((((....(((((((..(((((......)))))..)))))))...(..((((((((.((....)).-...((.(((((....))))).))..))))..))))..)..))))))..... ( -39.20) >DroYak_CAF1 82128 111 + 1 AGCAAUUCGAUUCGAUUUGGCUUCGGAUUUGAAUUUGGAUUGAGAAA--------UUGAGGAAUGCUA-UCUCAGCUGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU (((((((((((((((((..((....)..)..)).))))))))))...--------(..((((..(((.-....)))((((....))))............))))..)...)))))..... ( -28.80) >consensus AGCAAUUCGAUUCGAUUUGGUUUCGGAUUGGAAUUUGUAUUGAGGAA________UUGAGGAAUGCUA_UCUCCGGUGUGGUGACACACGUGCCCAAUUAUCCUUGGAUAUUGCUUUAAU (((((((((((((((.......)))))))))).......(..((((...........((((........)))).((((((......)))..)))......))))..)...)))))..... (-24.16 = -24.40 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:35 2006