| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,808,934 – 12,809,053 |
| Length | 119 |
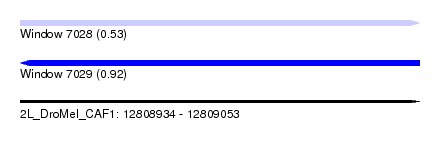
| Max. P | 0.922117 |

| Location | 12,808,934 – 12,809,053 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12808934 119 + 22407834 AGUUUGUCAAGGUUCGGGAGAUGGCAUGACAAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGAGGCCGCAAAAUGGUUCGGAGUCAAA- ..........(..((((((....(((((.((.(((((.......))))).))..)))))..)))).........(((..(.....((((.......))))....)..))).))..)...- ( -29.00) >DroSec_CAF1 74677 112 + 1 AGUUUGUCAAGGUUCGGGAGAUGGCAUGACAAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGAGGCCGC-------UUCGGAAGGAAA- ............(((((((....(((((.((.(((((.......))))).))..)))))..))))...............(.(((((((.......))))-------))).)...))).- ( -32.30) >DroSim_CAF1 77925 113 + 1 AGUUUGUCAAGGUUCGGGAGAUGGCAUGACAAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGAGGCCGC-------UUCGGAAGGAAAA ............(((((((....(((((.((.(((((.......))))).))..)))))..))))...............(.(((((((.......))))-------))).)...))).. ( -32.30) >DroEre_CAF1 78051 118 + 1 AGUUUGUCAGGGCUCG-AAGAUGGCAUGACUAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGAGGGCGCAAAAUGGCUGGGUAAUGAA- .....((((.(((((.-......(((((.((((((((.......))))).))).)))))..(((....)))...)))))......(((.........)))....))))...........- ( -26.30) >DroYak_CAF1 78247 119 + 1 AGUUUGUCAGGGCUCGGGAGAUGGCAUGACAAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGCGGCAGCAAAAUGCUUUGGUAAUGAA- ......((....((((((.(((.(((((.((.(((((.......))))).))..))))).))))).))))....))(((((.((((((....((....))....)))))).)))))...- ( -35.90) >consensus AGUUUGUCAAGGUUCGGGAGAUGGCAUGACAAGAUAUAAAGUCCAUAUCAUGGACAUGCAAUCCCACGGGAAAAGAGUUACGGAAGCGGAAAGAGGCCGCAAAAUG_UUCGGAAAGAAA_ ..(((((((.(.(((....)).).).))))))).......((((((...))))))..((..(((....))).........((....))..........)).................... (-21.88 = -21.84 + -0.04)



| Location | 12,808,934 – 12,809,053 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
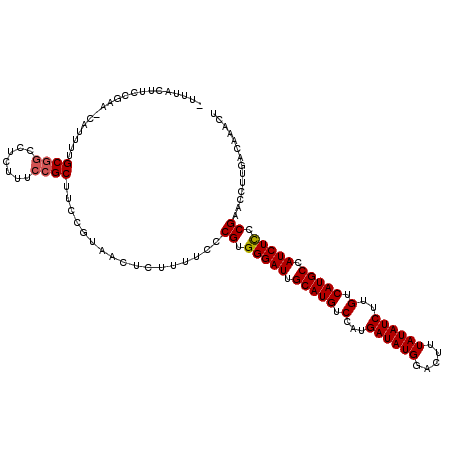
>2L_DroMel_CAF1 12808934 119 - 22407834 -UUUGACUCCGAACCAUUUUGCGGCCUCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUUGUCAUGCCAUCUCCCGAACCUUGACAAACU -((((...............((((.......)))).................((.(((((.(((((..((.((((((.....)))))).)).))))).))))).))........)))).. ( -24.40) >DroSec_CAF1 74677 112 - 1 -UUUCCUUCCGAA-------GCGGCCUCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUUGUCAUGCCAUCUCCCGAACCUUGACAAACU -.........(((-------((((.......)))))))..............((.(((((.(((((..((.((((((.....)))))).)).))))).))))).)).............. ( -28.80) >DroSim_CAF1 77925 113 - 1 UUUUCCUUCCGAA-------GCGGCCUCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUUGUCAUGCCAUCUCCCGAACCUUGACAAACU ..........(((-------((((.......)))))))..............((.(((((.(((((..((.((((((.....)))))).)).))))).))))).)).............. ( -28.80) >DroEre_CAF1 78051 118 - 1 -UUCAUUACCCAGCCAUUUUGCGCCCUCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUAGUCAUGCCAUCUU-CGAGCCCUGACAAACU -................((((((..(((...((((..................))))(((.(((((.(...((((((.....))))))..).))))).)))..-.)))...)).)))).. ( -19.07) >DroYak_CAF1 78247 119 - 1 -UUCAUUACCAAAGCAUUUUGCUGCCGCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUUGUCAUGCCAUCUCCCGAGCCCUGACAAACU -.(((((((..((((.....((....)).....))))..)))).........((.(((((.(((((..((.((((((.....)))))).)).))))).))))).)).....)))...... ( -25.60) >consensus _UUUACUUCCGAA_CAUUUUGCGGCCUCUUUCCGCUUCCGUAACUCUUUUCCCGUGGGAUUGCAUGUCCAUGAUAUGGACUUUAUAUCUUGUCAUGCCAUCUCCCGAACCUUGACAAACU ....................((((.......)))).................((.(((((.(((((.(...((((((.....))))))..).))))).))))).)).............. (-20.84 = -21.08 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:31 2006