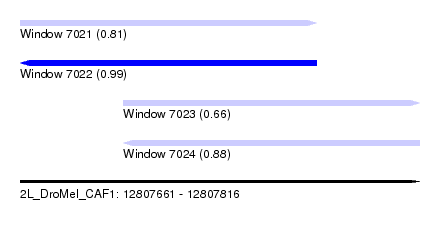
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,807,661 – 12,807,816 |
| Length | 155 |
| Max. P | 0.987408 |

| Location | 12,807,661 – 12,807,776 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -43.34 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807661 115 + 22407834 UUUUUUUACUUUCUGGUCUUUUAAGAGCCCAGUUGAGUGGCUCACCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGUUCUUGCCACCG-----CCGCUCAUCGUUAGCCA .......((((.((((.(((....))).))))..))))((((.((..(((((((((((((((..........)))))))))(.((((....)))).))-----)).)))...)).)))). ( -40.60) >DroSec_CAF1 73420 115 + 1 UUUUUUUACUUUCUGGUCUUUUAAGAGCCCAGUUGAGUGGCUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCA ............((((.(((....))).)))).((((((((..(((....)))(((((((((..........)))))))))(.((((....)))).))-----))))))).......... ( -43.50) >DroSim_CAF1 76666 112 + 1 UUUUU---CUUUUUGGUCUUUUAAGAGCCCAGUUGAGUGGCUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCA .....---.....(((.(((....))).)))(.((((((((..(((....)))(((((((((..........)))))))))(.((((....)))).))-----))))))).)........ ( -41.80) >DroEre_CAF1 76777 111 + 1 ----UUUACUUUCUGGUCCUUUAAGAGCCCGGUUGAGUGGCUCGCCCGGGGGCGCGAUGUUGUCGUGGGGCUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCA ----..........((..(.....)..))((((.(((((((..(((....)))(((((((((((....)))..))))))))(.((((....)))).))-----))))))))))....... ( -44.00) >DroYak_CAF1 76953 116 + 1 ----UUUACUUUCUGGUCUUUUAAGAGCCCAGUUGAGUGGCUUGCCCGGGGGCGCAAAGUUGUCGUAGGGCUUGACAUCGCGAUGGCUCUUCCCACCGCGGCGCCGCUCGUCGUUAGCCA ----...((((.((((.(((....))).))))..))))((((.((.(((((((((.....(((((.......))))).((((.(((......))).))))))))).))))..)).)))). ( -46.80) >consensus UUUUUUUACUUUCUGGUCUUUUAAGAGCCCAGUUGAGUGGCUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG_____CCGCUCAUCGUUAGCCA ............((((.(((....))).)))).(((((((......(((.((((.((.((..(((((((......).))))))..)))).)))).))).....))))))).......... (-36.00 = -36.16 + 0.16)

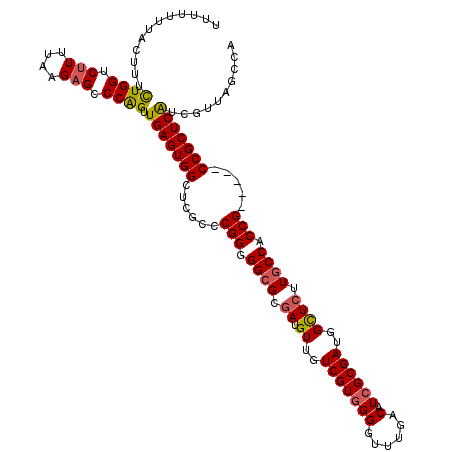
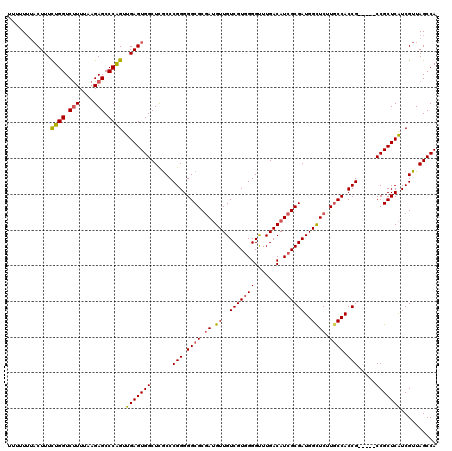
| Location | 12,807,661 – 12,807,776 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -32.28 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807661 115 - 22407834 UGGCUAACGAUGAGCGG-----CGGUGGCAAGAACCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGUGAGCCACUCAACUGGGCUCUUAAAAGACCAGAAAGUAAAAAAA (((((..(((((....(-----((((((......)))))))..((((.........)))).)))))((((....))))))))).....((((.((......)).))))............ ( -38.10) >DroSec_CAF1 73420 115 - 1 UGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAGCCACUCAACUGGGCUCUUAAAAGACCAGAAAGUAAAAAAA (((((..(((((....(-----(((((((....))))))))..((((.........)))).)))))((((....))))))))).....((((.((......)).))))............ ( -43.60) >DroSim_CAF1 76666 112 - 1 UGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAGCCACUCAACUGGGCUCUUAAAAGACCAAAAAG---AAAAA (((((..(((((....(-----(((((((....))))))))..((((.........)))).))))).(((....)))(((((.(......)))))).....)).))).....---..... ( -41.10) >DroEre_CAF1 76777 111 - 1 UGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAGCCCCACGACAACAUCGCGCCCCCGGGCGAGCCACUCAACCGGGCUCUUAAAGGACCAGAAAGUAAA---- (((((......((((((-----(((((((....))))))(((((((..............)))))))))).((((..(((...)))..)))))))).....)).))).........---- ( -42.34) >DroYak_CAF1 76953 116 - 1 UGGCUAACGACGAGCGGCGCCGCGGUGGGAAGAGCCAUCGCGAUGUCAAGCCCUACGACAACUUUGCGCCCCCGGGCAAGCCACUCAACUGGGCUCUUAAAAGACCAGAAAGUAAA---- ..(((......(((.(((((((((((((......)))))))).((((.........)))).....))(((....)))..))).)))..((((.((......)).))))..)))...---- ( -41.50) >consensus UGGCUAACGAUGAGCGG_____CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAGCCACUCAACUGGGCUCUUAAAAGACCAGAAAGUAAAAAAA (((((..(((((..........(((((((....)))))))...((((.........)))).)))))((((....))))))))).....((((.((......)).))))............ (-32.28 = -33.00 + 0.72)

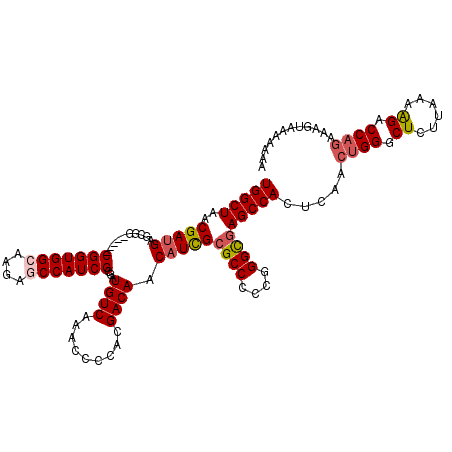
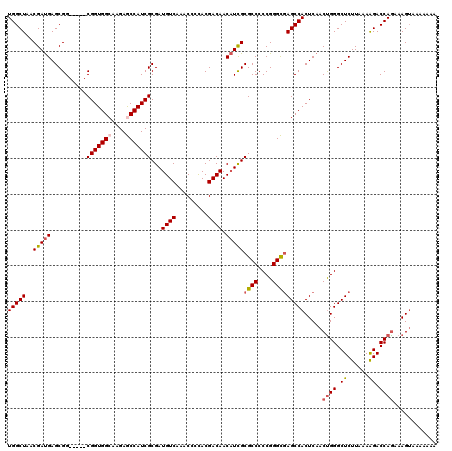
| Location | 12,807,701 – 12,807,816 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -47.44 |
| Consensus MFE | -39.81 |
| Energy contribution | -39.85 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807701 115 + 22407834 CUCACCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGUUCUUGCCACCG-----CCGCUCAUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCUUUCCAC ....(((((((((((((((..((.(((((((..(((((....)))..))..))).)))-----).)).))))))..))).......(((((.........)))))))))))......... ( -45.80) >DroSec_CAF1 73460 115 + 1 CUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCAUUCCAC ...((((.(((((((((((.(((((.......)))))..(((.((((....)))).))-----)....))))))..)))......((((((.........)))))))).))))....... ( -48.30) >DroSim_CAF1 76703 115 + 1 CUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCAUUCCAC ...((((.(((((((((((.(((((.......)))))..(((.((((....)))).))-----)....))))))..)))......((((((.........)))))))).))))....... ( -48.30) >DroEre_CAF1 76813 115 + 1 CUCGCCCGGGGGCGCGAUGUUGUCGUGGGGCUUGACAUCGCGAUGGCUCUUGCCACCG-----CCGCUCAUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCCUUCCAC ...((((.(((((((((((..((.(((((((..(((((....)))..))..))).)))-----).)).))))))..)))......((((((.........)))))))).))))....... ( -48.60) >DroYak_CAF1 76989 120 + 1 CUUGCCCGGGGGCGCAAAGUUGUCGUAGGGCUUGACAUCGCGAUGGCUCUUCCCACCGCGGCGCCGCUCGUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCUUUCCAC ......(((((((((.....(((((.......))))).((((.(((......))).))))))))).))))..(..((((.((...((((((.........))))))..))))))..)... ( -46.20) >consensus CUCGCCCGGGGGCGCGAUGUUGUCGUGGGGUUUGACAUCGCGAUGGCUCUUGCCACCG_____CCGCUCAUCGUUAGCCACCUUAGUGUCCAAUAUGCCAGGACACCCGGGGCAUUCCAC ...((((.((((((((((((((((....)))..))))))))((((((..................)).))))....)))......((((((.........)))))))).))))....... (-39.81 = -39.85 + 0.04)

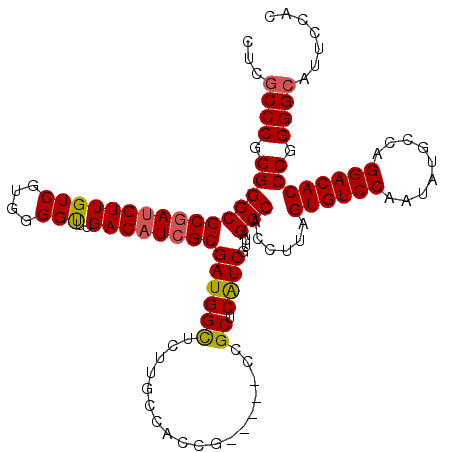
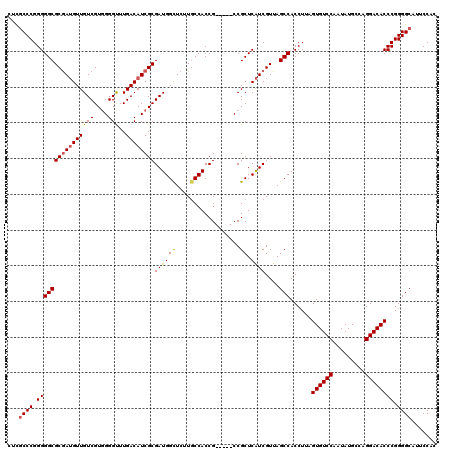
| Location | 12,807,701 – 12,807,816 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -48.94 |
| Consensus MFE | -41.86 |
| Energy contribution | -42.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807701 115 - 22407834 GUGGAAAGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGAUGAGCGG-----CGGUGGCAAGAACCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGUGAG ......((((((.(((((((.........)))))))..)).))))..(((((....(-----((((((......)))))))..((((.........)))).)))))((((....)))).. ( -45.10) >DroSec_CAF1 73460 115 - 1 GUGGAAUGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAG ......(((((..(((((((.........)))))))..((.(((...(((((....(-----(((((((....))))))))..((((.........)))).))))).))).))))))).. ( -50.20) >DroSim_CAF1 76703 115 - 1 GUGGAAUGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAG ......(((((..(((((((.........)))))))..((.(((...(((((....(-----(((((((....))))))))..((((.........)))).))))).))).))))))).. ( -50.20) >DroEre_CAF1 76813 115 - 1 GUGGAAGGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGAUGAGCGG-----CGGUGGCAAGAGCCAUCGCGAUGUCAAGCCCCACGACAACAUCGCGCCCCCGGGCGAG ......((((((.(((((((.........)))))))..)).))))..(((((....(-----(((((((....))))))))..((((.........)))).)))))((((....)))).. ( -50.60) >DroYak_CAF1 76989 120 - 1 GUGGAAAGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGACGAGCGGCGCCGCGGUGGGAAGAGCCAUCGCGAUGUCAAGCCCUACGACAACUUUGCGCCCCCGGGCAAG .......((((..(((((((.........)))))))..((.((((.......)))(((((((((((((......)))))))).((((.........)))).....))))))))))))... ( -48.60) >consensus GUGGAAAGCCCCGGGUGUCCUGGCAUAUUGGACACUAAGGUGGCUAACGAUGAGCGG_____CGGUGGCAAGAGCCAUCGCGAUGUCAAACCCCACGACAACAUCGCGCCCCCGGGCGAG .......((((..(((((((.........)))))))..((.(((...................((((((....))))))(((((((..............)))))))))).))))))... (-41.86 = -42.14 + 0.28)

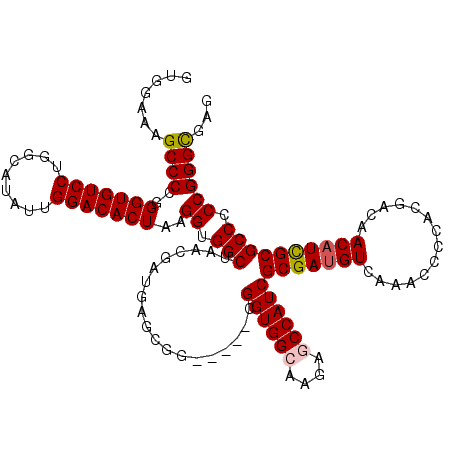
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:26 2006