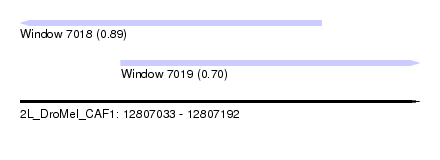
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,807,033 – 12,807,192 |
| Length | 159 |
| Max. P | 0.894211 |

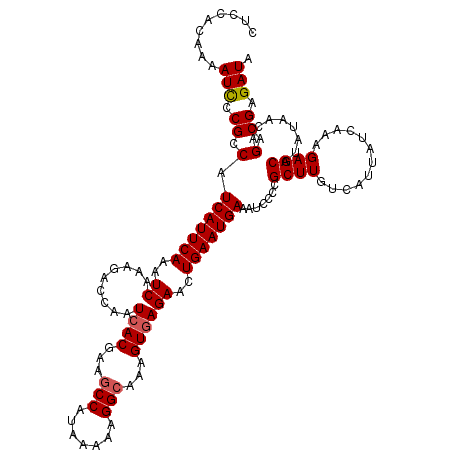
| Location | 12,807,033 – 12,807,153 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807033 120 - 22407834 CUCCACAAAAUCCCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAGCCAUAAAAAGGCAAAGUGAGAACUGAAUGAAAUCCCCGCUUGUCAUUAUCAAAGAGCAUAUAACAGACGAGAUA .........(((.((.(.(((((((..((.........((((...(((.......)))...))))))..))))))).......((((............))))........).)).))). ( -20.10) >DroSec_CAF1 72723 120 - 1 CUCCACAAAAUCCCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAACCAUGAAAAGGCAAAGUCAGAACUGAAUGAAAUCCCCGCUUGUCAUUAUCAAAGAGCAUAUAACAGGCGAGAUA .........(((.((((.(((((((..((.........(((.(....).)))...(((...))).))..))))))).......((((............))))........)))).))). ( -21.80) >DroSim_CAF1 75994 120 - 1 CUCCACAAAAUCCCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAGCCAUGAAAAGGCAAAGUGAGAACUGAAUGAAAUCCCCGCUUGUCAUUAUCAAAGAGCAUAUAACAGACGAGAUA .........(((.((.(.(((((((..((.........((((...(((.......)))...))))))..))))))).......((((............))))........).)).))). ( -20.10) >DroEre_CAF1 76174 119 - 1 CCCCACAAAAUUUCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAGCCAUAAAAAGGCAAAGUGAGAACUGAAUGAAAUC-CCGCUUGUCAUUAUCAAAGAGCGUAUAACAGACGAGAUA .........((((((.(.(((((((..((.........((((...(((.......)))...))))))..)))))))....-.(((((............))))).......).)))))). ( -23.60) >DroYak_CAF1 76039 120 - 1 CUUCACGAAAUCCCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAGCCAUAAAAAGGCAAAGUGAGAACUGAAUGAAAUCCCCGCUUGUCAUUAUCAAAGAGCAUAUAACAGACGAGAUA .........(((.((.(.(((((((..((.........((((...(((.......)))...))))))..))))))).......((((............))))........).)).))). ( -20.10) >consensus CUCCACAAAAUCCCGCCAUCAUUCAAAUCAAAGACCAAUCACGAAGCCAUAAAAAGGCAAAGUGAGAACUGAAUGAAAUCCCCGCUUGUCAUUAUCAAAGAGCAUAUAACAGACGAGAUA .........(((.((.(.(((((((..((.........((((...(((.......)))...))))))..))))))).......((((............))))........).)).))). (-17.72 = -17.96 + 0.24)

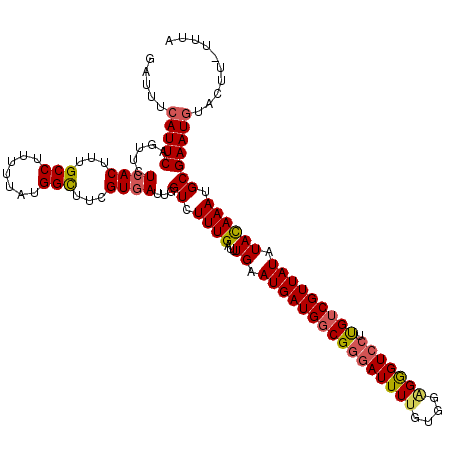
| Location | 12,807,073 – 12,807,192 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12807073 119 + 22407834 GAUUUCAUUCAGUUCUCACUUUGCCUUUUUAUGGCUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGGGAUUUUGUGGAGGGUCCUUGUCGUUAUAUACAAAUGCGAAUGUACUU-UUUA .....(((((.....((((...(((.......)))...))))..........(((((.((((((((((((((((....))))))).)))))))))...)))))..))))).....-.... ( -28.90) >DroSec_CAF1 72763 120 + 1 GAUUUCAUUCAGUUCUGACUUUGCCUUUUCAUGGUUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGGGAUUUUGUGGAGGGUCCUUGUCGUUAUAUACAAAUGCGAAUGUACUUUUUUA .....(((((............(((...(((((....)))))..))).....(((((.((((((((((((((((....))))))).)))))))))...)))))..))))).......... ( -27.40) >DroSim_CAF1 76034 120 + 1 GAUUUCAUUCAGUUCUCACUUUGCCUUUUCAUGGCUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGGGAUUUUGUGGAGGGUCCUUGACGUUAUAUACAAAUGCGAAUGUACUUUUUUA .....(((((.....((((...(((.......)))...))))..........(((((.((((((..((((((((....))))))))...))))))...)))))..))))).......... ( -25.40) >DroEre_CAF1 76213 119 + 1 GAUUUCAUUCAGUUCUCACUUUGCCUUUUUAUGGCUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGAAAUUUUGUGGGGAGUGCUCGUCGUUAUAUAUAAAUGCGAAUGUACUU-GUUA ..............(((((...(((.......)))(((((.(((..((........))..))).))))).....)))))((((((...((((...........))))..))))))-.... ( -28.30) >DroYak_CAF1 76079 119 + 1 GAUUUCAUUCAGUUCUCACUUUGCCUUUUUAUGGCUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGGGAUUUCGUGAAGAGUCCUCGUCGUUAUAUACAAAGGCGAAUAUACUA-UUAA ......((((.....((((...(((.......)))...))))...(((((((...((.(((((((((((((((......)))))).))))))))).)))))))))))))......-.... ( -33.90) >consensus GAUUUCAUUCAGUUCUCACUUUGCCUUUUUAUGGCUUCGUGAUUGGUCUUUGAUUUGAAUGAUGGCGGGAUUUUGUGGAGGGUCCUUGUCGUUAUAUACAAAUGCGAAUGUACUU_UUUA .....(((((.....((((...(((.......)))...))))...((.((((...((.((((((((((((((((....))))))).))))))))).)))))).))))))).......... (-24.50 = -24.78 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:22 2006