
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,805,072 – 12,805,262 |
| Length | 190 |
| Max. P | 0.999455 |

| Location | 12,805,072 – 12,805,192 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -38.50 |
| Energy contribution | -38.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12805072 120 + 22407834 GGAUGCGGAUGAUCGGAAGGGGGAUCGGGCUAAGGACGAUGGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUG ....((...(((((........))))).)).................(((((.(((..(((((......)))))......(((((.(((((((.....))))))).)))))))).))))) ( -40.30) >DroSec_CAF1 70831 120 + 1 GGAUGCGGAUGAUCGGGAGGGGGAUCGGACUAAGGACGAUGGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUG ......(..(((((........)))))..).................(((((.(((..(((((......)))))......(((((.(((((((.....))))))).)))))))).))))) ( -40.60) >DroEre_CAF1 74208 120 + 1 GGAUGUGGAUGAUCAGGAGGGGGAUCGGACUAAGGACGUCGGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUG .((((((..(((((........)))))..).....))))).......(((((.(((..(((((......)))))......(((((.(((((((.....))))))).)))))))).))))) ( -43.50) >DroYak_CAF1 74080 119 + 1 -GAUGCGGAUGAUCAGUAGGGGGAUCGGAGCAAGGACGAUGGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUG -..(((...(((((........)))))..)))...............(((((.(((..(((((......)))))......(((((.(((((((.....))))))).)))))))).))))) ( -43.10) >consensus GGAUGCGGAUGAUCAGGAGGGGGAUCGGACUAAGGACGAUGGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUG .........(((((........)))))....................(((((.(((..(((((......)))))......(((((.(((((((.....))))))).)))))))).))))) (-38.50 = -38.50 + 0.00)

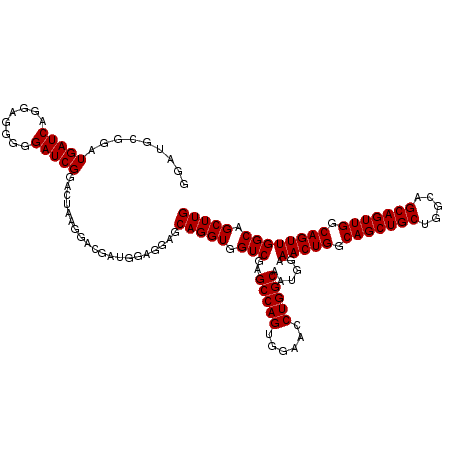
| Location | 12,805,072 – 12,805,192 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
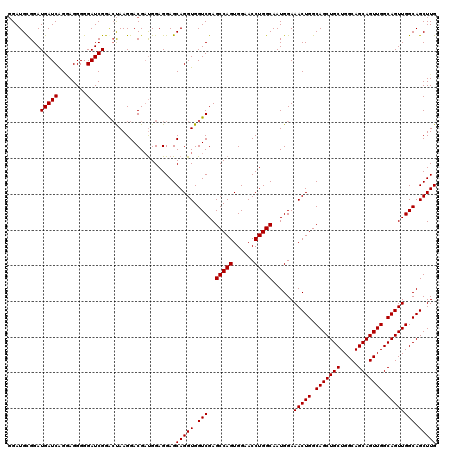
>2L_DroMel_CAF1 12805072 120 - 22407834 CAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUUUCCAUUGCCAGGUUCCACUGGCUCGACCACCUGCUCCUCCAUCGUCCUUAGCCCGAUCCCCCUUCCGAUCAUCCGCAUCC ...((((..(((((.((.((((.....)))).)).)))))......(((((......)))))..........................))))..((((........)))).......... ( -26.90) >DroSec_CAF1 70831 120 - 1 CAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUUUCCAUUGCCAGGUUCCACUGGCUCGACCACCUGCUCCUCCAUCGUCCUUAGUCCGAUCCCCCUCCCGAUCAUCCGCAUCC ...((.((.(((((.((.((((.....)))).)).)))))......(((((......)))))..........))....................((((........))))....)).... ( -26.80) >DroEre_CAF1 74208 120 - 1 CAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUUUCCAUUGCCAGGUUCCACUGGCUCGACCACCUGCUCCUCCGACGUCCUUAGUCCGAUCCCCCUCCUGAUCAUCCACAUCC ..(((((..(((((.((.((((.....)))).)).)))))..))..(((((......)))))..........))).....(((.......))).((((........)))).......... ( -28.20) >DroYak_CAF1 74080 119 - 1 CAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUUUCCAUUGCCAGGUUCCACUGGCUCGACCACCUGCUCCUCCAUCGUCCUUGCUCCGAUCCCCCUACUGAUCAUCCGCAUC- ..(((((..(((((.((.((((.....)))).)).)))))..))..(((((......)))))..........))).............(((...((((........))))....)))..- ( -27.30) >consensus CAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUUUCCAUUGCCAGGUUCCACUGGCUCGACCACCUGCUCCUCCAUCGUCCUUAGUCCGAUCCCCCUCCCGAUCAUCCGCAUCC ..(((((..(((((.((.((((.....)))).)).)))))..))..(((((......)))))..........)))...................((((........)))).......... (-26.25 = -26.25 + 0.00)


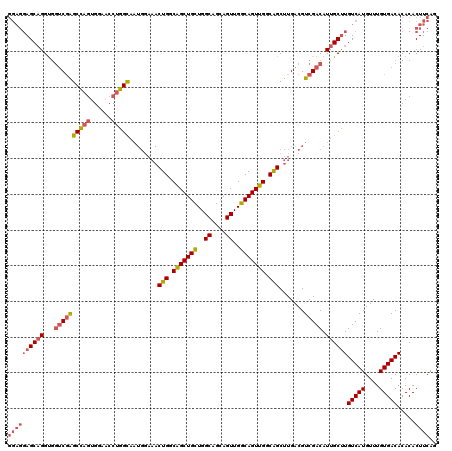
| Location | 12,805,112 – 12,805,232 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -52.22 |
| Consensus MFE | -43.02 |
| Energy contribution | -44.46 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
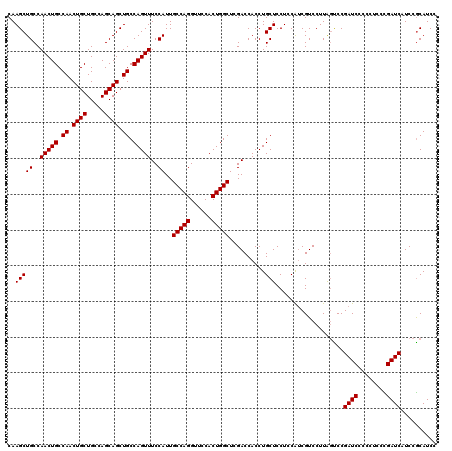
>2L_DroMel_CAF1 12805112 120 + 22407834 GGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAG ((((((((((...((((((((((......)))))..((..(((((.(((((((..((....))..))))))).))).))..))))))).))))))(((((....))))).....)))).. ( -54.30) >DroPse_CAF1 99042 104 + 1 -----AGCUGGCUAACAGGCUCCUGGAGCCUGGU--------CUGGCGGCUGUUGGC---AGUUGGCAGUUGCCGGCUUGACGUCGACAUUGCCUGUCAUGUUUGUGACACACACUUCAG -----.((..(....(((((((...)))))))..--------)..))((((((((((---(((((((....)))))))....)))))))..)))((((((....)))))).......... ( -43.90) >DroSec_CAF1 70871 120 + 1 GGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAG ((((((((((...((((((((((......)))))..((..(((((.(((((((..((....))..))))))).))).))..))))))).))))))(((((....))))).....)))).. ( -54.30) >DroEre_CAF1 74248 120 + 1 GGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAG ((((((((((...((((((((((......)))))..((..(((((.(((((((..((....))..))))))).))).))..))))))).))))))(((((....))))).....)))).. ( -54.30) >DroYak_CAF1 74119 120 + 1 GGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAG ((((((((((...((((((((((......)))))..((..(((((.(((((((..((....))..))))))).))).))..))))))).))))))(((((....))))).....)))).. ( -54.30) >consensus GGAGGAGCAGGUGGUCGAGCCAGUGGAACCUGGCAAUGGAAACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAG ((((((((((...((((((((((......)))))........(((.(((((((..((....))..))))))).))).......))))).))))))(((((....))))).....)))).. (-43.02 = -44.46 + 1.44)

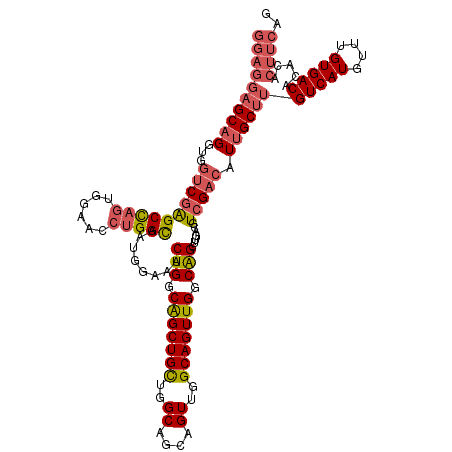
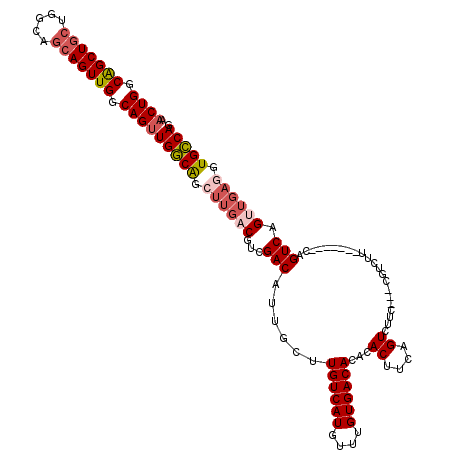
| Location | 12,805,152 – 12,805,262 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -33.14 |
| Energy contribution | -34.50 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12805152 110 + 22407834 AACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAGUCUUC---CGUCUU-------CAGUCAGUUGAGGUGCCAG (((((.(((((((.....))))))).)))))((((.((..((...(((..((..((((((....))))))..))....((.(...---.).)).-------..))).))..)).)))).. ( -42.00) >DroPse_CAF1 99071 103 + 1 --CUGGCGGCUGUUGGC---AGUUGGCAGUUGCCGGCUUGACGUCGACAUUGCCUGUCAUGUUUGUGACACACACUUCAGUCCUCGGCCGUCUU-------CAGUCAG-----GUGGCAG --..((((((((..(((---....((((((.(.((((.....)))).)))))))((((((....)))))).........)))..))))))))..-------..(((..-----..))).. ( -38.50) >DroSec_CAF1 70911 103 + 1 AACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAGUC----------UU-------CAGUCAGCUGAAGUGCCAG (((((.(((((((.....))))))).)))))(((.((..(((((.((((.....))))))))).)).......(((((((.(----------(.-------.....)))))))))))).. ( -40.50) >DroEre_CAF1 74288 110 + 1 AACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAGUCUUC---CGUCUU-------CAGUCAGUUGAGGUGCCAG (((((.(((((((.....))))))).)))))((((.((..((...(((..((..((((((....))))))..))....((.(...---.).)).-------..))).))..)).)))).. ( -42.00) >DroYak_CAF1 74159 117 + 1 AACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAGUCUUC---CGUCUUCAGUCUUCAGUCAGUUGAGGUGCCAG (((((.(((((((.....))))))).)))))((((.((..((...(((......((((((....))))))...(((..((.(...---.).))..))).....))).))..)).)))).. ( -43.00) >consensus AACUGGCAGCUGCUGGCAGCAGUUGGCAGUUGGCAGCUUGACGUCGACAUUGCUUGUCAUGUUUGUGACACACACUUCAGUCUUC___CGUCUU_______CAGUCAGUUGAGGUGCCAG .((((.(((((((.....))))))).))))(((((.((((((...(((......((((((....))))))...((....))......................))).)))))).))))). (-33.14 = -34.50 + 1.36)

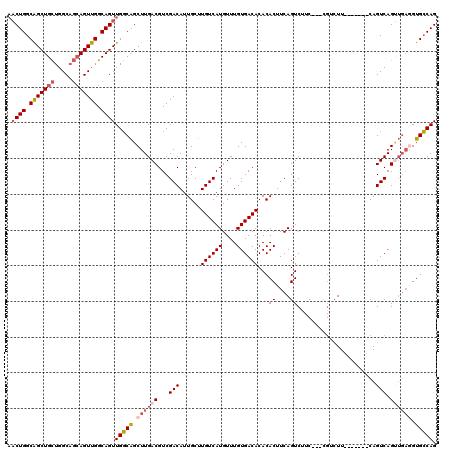
| Location | 12,805,152 – 12,805,262 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -25.16 |
| Energy contribution | -26.36 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12805152 110 - 22407834 CUGGCACCUCAACUGACUG-------AAGACG---GAAGACUGAAGUGUGUGUCACAAACAUGACAAGCAAUGUCGACGUCAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUU (((((..............-------..((((---...(((.....(((.(((((......))))).)))..)))..)))).((((((.....((.....)).....))))))))))).. ( -36.20) >DroPse_CAF1 99071 103 - 1 CUGCCAC-----CUGACUG-------AAGACGGCCGAGGACUGAAGUGUGUGUCACAAACAUGACAGGCAAUGUCGACGUCAAGCCGGCAACUGCCAACU---GCCAACAGCCGCCAG-- .((((..-----((.....-------.))..(((....((((((..(((.(((((......))))).)))...)))..)))..))))))).(((.(..((---(....)))..).)))-- ( -27.50) >DroSec_CAF1 70911 103 - 1 CUGGCACUUCAGCUGACUG-------AA----------GACUGAAGUGUGUGUCACAAACAUGACAAGCAAUGUCGACGUCAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUU ((((((...((((((((.(-------..----------(((.....(((.(((((......))))).)))..)))..)))).))))).............((((....)))))))))).. ( -35.00) >DroEre_CAF1 74288 110 - 1 CUGGCACCUCAACUGACUG-------AAGACG---GAAGACUGAAGUGUGUGUCACAAACAUGACAAGCAAUGUCGACGUCAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUU (((((..............-------..((((---...(((.....(((.(((((......))))).)))..)))..)))).((((((.....((.....)).....))))))))))).. ( -36.20) >DroYak_CAF1 74159 117 - 1 CUGGCACCUCAACUGACUGAAGACUGAAGACG---GAAGACUGAAGUGUGUGUCACAAACAUGACAAGCAAUGUCGACGUCAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUU (((((.......................((((---...(((.....(((.(((((......))))).)))..)))..)))).((((((.....((.....)).....))))))))))).. ( -36.20) >consensus CUGGCACCUCAACUGACUG_______AAGACG___GAAGACUGAAGUGUGUGUCACAAACAUGACAAGCAAUGUCGACGUCAAGCUGCCAACUGCCAACUGCUGCCAGCAGCUGCCAGUU (((((.................................((((((..(((.(((((......))))).)))...)))..))).((((((.....((.....)).....))))))))))).. (-25.16 = -26.36 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:20 2006