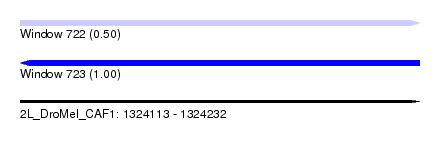
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,324,113 – 1,324,232 |
| Length | 119 |
| Max. P | 0.997548 |

| Location | 1,324,113 – 1,324,232 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -29.95 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1324113 119 + 22407834 G-UUUUGUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCAAAAGCCGAGAUAAAAACUAAGGCAA .-((((((.....))))))....(((((((((((...(((....)))...))))))))..(((..((((.((((((....)))))).))))(((....))))))...........))).. ( -31.60) >DroSec_CAF1 81529 119 + 1 G-UUUUGUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCAAAAGCCGAGAUAAAAACUAAGGCAA .-((((((.....))))))....(((((((((((...(((....)))...))))))))..(((..((((.((((((....)))))).))))(((....))))))...........))).. ( -31.60) >DroSim_CAF1 85017 119 + 1 G-UUUUGUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCAAAAGCCGAGAUAAAAACUAAGGCAA .-((((((.....))))))....(((((((((((...(((....)))...))))))))..(((..((((.((((((....)))))).))))(((....))))))...........))).. ( -31.60) >DroEre_CAF1 94691 117 + 1 G---UUUUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGCGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCCAAAGCCGAGAUAAAAACUAAGGCAA .---(((((.....)))))..((((((((..((((....)))).((((((.(.(((((((..((...)).))))))).).))))))..))))))))..(((..............))).. ( -34.74) >DroYak_CAF1 84791 119 + 1 G-GUUUUUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCAAAAGCCGAGAUAAAAACUAAGGCAA (-(((((((....(((((.....((((((..((((((((................)))))))).))))))((((((....)))))))))))(((....)))....))))))))....... ( -31.89) >DroAna_CAF1 107801 119 + 1 GAGGUUUUGGAAGACAAAGUACGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGCUUGCAUUUGAAUGCAAUUUGUGGCA-AAGCCGAGAUAAAAACUAAGGCAA ...(((((((...(((((....(((((((..((((((((................)))))))).)))))))(((((....))))).)))))(((.-..)))..........))))))).. ( -33.69) >consensus G_UUUUGUGGAAGACAAAGUAUGGCCGCAUUGGGUUACGACUCCUUGCAUCCUAAUGUGACUCCUGCGGUUUGCAUUUGAAUGCAAUUUGUGGCAAAAGCCGAGAUAAAAACUAAGGCAA .......................(((((((((((...(((....)))...))))))))..(((..((((.((((((....)))))).))))(((....))))))...........))).. (-29.95 = -29.82 + -0.14)

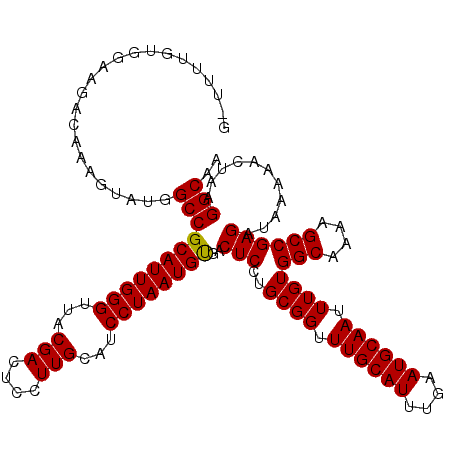
| Location | 1,324,113 – 1,324,232 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.75 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1324113 119 - 22407834 UUGCCUUAGUUUUUAUCUCGGCUUUUGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCACAAAA-C ........(((((......(((((((((......((((((....))))))...)))))))))......(((.((((((.((((((......))))))....))))))..)))..))))-) ( -30.60) >DroSec_CAF1 81529 119 - 1 UUGCCUUAGUUUUUAUCUCGGCUUUUGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCACAAAA-C ........(((((......(((((((((......((((((....))))))...)))))))))......(((.((((((.((((((......))))))....))))))..)))..))))-) ( -30.60) >DroSim_CAF1 85017 119 - 1 UUGCCUUAGUUUUUAUCUCGGCUUUUGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCACAAAA-C ........(((((......(((((((((......((((((....))))))...)))))))))......(((.((((((.((((((......))))))....))))))..)))..))))-) ( -30.60) >DroEre_CAF1 94691 117 - 1 UUGCCUUAGUUUUUAUCUCGGCUUUGGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCGCAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCAAAA---C ..(((..((((........))))..))).......(((((((...(((.....))).)))).)))...(((.((((((.((((((......))))))....))))))..)))....---. ( -30.00) >DroYak_CAF1 84791 119 - 1 UUGCCUUAGUUUUUAUCUCGGCUUUUGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCAAAAAC-C ........((((((.....(((((((((......((((((....))))))...)))))))))......(((.((((((.((((((......))))))....))))))..)))))))))-. ( -32.90) >DroAna_CAF1 107801 119 - 1 UUGCCUUAGUUUUUAUCUCGGCUU-UGCCACAAAUUGCAUUCAAAUGCAAGCCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCGUACUUUGUCUUCCAAAACCUC ........(((((......(((((-(((......((((((....))))))...))).)))))......(((.((((((.((((((......))))))....))))))..))))))))... ( -28.20) >consensus UUGCCUUAGUUUUUAUCUCGGCUUUUGCCACAAAUUGCAUUCAAAUGCAAACCGCAGGAGUCACAUUAGGAUGCAAGGAGUCGUAACCCAAUGCGGCCAUACUUUGUCUUCCAAAAAA_C ...................(((((((((......((((((....))))))...)))))))))......(((.((((((.((((((......))))))....))))))..)))........ (-29.42 = -29.75 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:25 2006