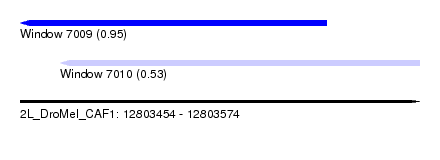
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,803,454 – 12,803,574 |
| Length | 120 |
| Max. P | 0.953820 |

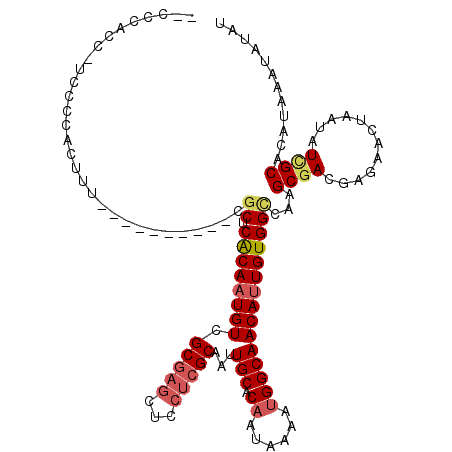
| Location | 12,803,454 – 12,803,546 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.46 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12803454 92 - 22407834 UCGCGAGCUUCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGUCAAGCGACGAGAACUAAUAUAGCACAUAAAUAUAUAUAUGUAUGUAU ..((.((.(((((((..(((..((....((((....)))).))..)))..).))))))))......))(((((.(((....))).))))).. ( -19.70) >DroSec_CAF1 69147 92 - 1 UCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAUAUAUGUAUGUAU ..((((.(((.((((....((.((....((((....)))).))))...)))).)))........))))(((((.(((....))).))))).. ( -23.70) >DroSim_CAF1 71309 92 - 1 UCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAUAUAUGUAUGUAU ..((((.(((.((((....((.((....((((....)))).))))...)))).)))........))))(((((.(((....))).))))).. ( -23.70) >DroYak_CAF1 72050 92 - 1 UCGCGAGCUCCUCGCGAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUUGCACAUAAAUAUAUAUAUGUAUGUAU ..((((.(((.((((....((.((....((((....)))).))))...)))).)))........))))(((((.(((....))).))))).. ( -22.70) >consensus UCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAUAUAUGUAUGUAU ..((((.(((.((((....((.((....((((....)))).))))...)))).)))........))))(((((.(((....))).))))).. (-21.67 = -21.43 + -0.25)

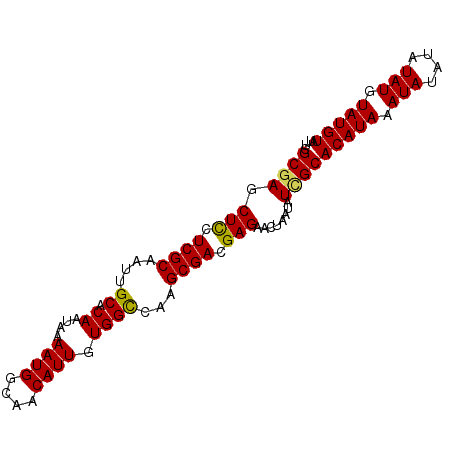
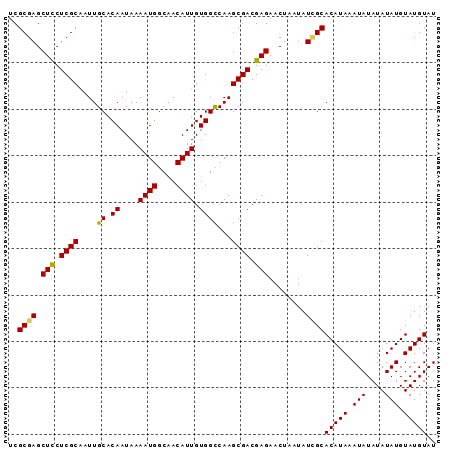
| Location | 12,803,466 – 12,803,574 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.17 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12803466 108 - 22407834 --CCCACAUUCCCCACUUU----------CGCUCACAAUGUCGCGAGCUUCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGUCAAGCGACGAGAACUAAUAUAGCACAUAAAUAUAU --.............((((----------((((((((((((.(((((...)))))...(((.((......)))))))))))))....))))).)))........................ ( -25.10) >DroPse_CAF1 98173 117 - 1 --CCCUCC-CCUGCCCUUGUGCCAUGAAGCACUCGCAUUGUCGCGCCCUUUUCGCAAUUGCACAAUAAAACGGCAACAUUGUGGCCCAGCGACGAGGACUAAUAUCGCAUAUAAAUAUAC --.((((.-..(((....((((......))))..)))..((((((((......((....))(((((.....(....)))))))))...)))))))))....................... ( -27.40) >DroSec_CAF1 69159 107 - 1 --CCCACC-ACCCCACUUU----------CGCUCACAAUGUCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAU --......-......((((----------((((((((((((.(((((...)))))...(((.((......)))))))))))))....))))).)))........................ ( -25.10) >DroSim_CAF1 71321 107 - 1 --CCCACC-UCCCCACUUU----------CGCUCACAAUGUCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAU --......-......((((----------((((((((((((.(((((...)))))...(((.((......)))))))))))))....))))).)))........................ ( -25.10) >DroEre_CAF1 71251 104 - 1 ------CUUUCCCCACUUG----------GGCUCACAAUGUCGCCAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAU ------.............----------(((.((((((((.((((.......((....)).........)))).)))))))))))..((((............))))............ ( -27.79) >DroYak_CAF1 72062 110 - 1 UCCCAACUUUCCCCACUUU----------CGCUCACAAUGUCGCGAGCUCCUCGCGAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUUGCACAUAAAUAUAU ...............((((----------((((((((((((((((((...)))))))((((.((......)))))))))))))....))))).))).....(((((.......))))).. ( -28.30) >consensus __CCCACC_UCCCCACUUU__________CGCUCACAAUGUCGCGAGCUCCUCGCAAUUGCACAAUAAAAUGGCAACAUUGUGGCCAAGCGACGAGAACUAAUAUCGCACAUAAAUAUAU ..............................((.((((((((.(((((...)))))...(((.((......)))))))))))))))...((((............))))............ (-18.98 = -19.78 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:13 2006