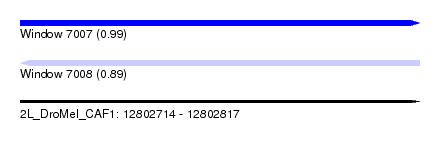
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,802,714 – 12,802,817 |
| Length | 103 |
| Max. P | 0.993189 |

| Location | 12,802,714 – 12,802,817 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12802714 103 + 22407834 CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCUGGUAAUGCGCCGCCUCUUCUCGCGGCCAAAAGGAUGCU--UC---------------CCCAUCCCCACAUACACUCCUUUAUUUU ....................(((((((....)))(((...((.(((((........)))))))...(((((..--..---------------..)))))))))))).............. ( -20.20) >DroPse_CAF1 96182 114 + 1 CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCUGGUAAUGUGCCGCCUCUCAUCGCUGCCAAACCCAUUCGCAUUUUAUUCCUGGCUUUGCCCACCGCCCCCCUCGCCCCUC------ .......................(((.....((((((...((.((.((........)).))))...)))...)))..........(((..........)))......)))....------ ( -15.40) >DroSec_CAF1 68345 103 + 1 CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCUGGUAAUGCGCCGCCUCUUCUCGCGGCCAAAAGGAUGCU--AC---------------CCCAUCCUCACAUAUACCCCUUUAUUUU .......................((((....))))((((.((.(((((........)))))))..((((((..--..---------------..))))))......)))).......... ( -22.40) >DroSim_CAF1 70522 103 + 1 CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCUGGUAAUGCGCCGCCUCUUCUCGCGGCCAAAAGGAUGCU--AC---------------CCCAUCCCCACAUAUACCCCUUUAUUUU .......................((((....))))((((.((.(((((........)))))))...(((((..--..---------------..))))).......)))).......... ( -21.40) >DroYak_CAF1 71296 106 + 1 CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCCGCUGAUGCGCCGCCUCUUCUCGCGGCCAAAAGGAUGCC--UCUCAU--------AAACCCACCCACCCACACACUCCUUU----U .......................((((((((((..((....))(((((........)))))))))))..))))--......--------..........................----. ( -20.40) >consensus CUCAUUUUAAUACACAUUUUUAUGGCACUUUUGCUGGUAAUGCGCCGCCUCUUCUCGCGGCCAAAAGGAUGCU__UC_______________CCCAUCCCCACAUACACCCCUUUAUUUU .......................((((....))))((((.((.(((((........)))))))...(((((.......................))))).......)))).......... (-13.52 = -14.08 + 0.56)

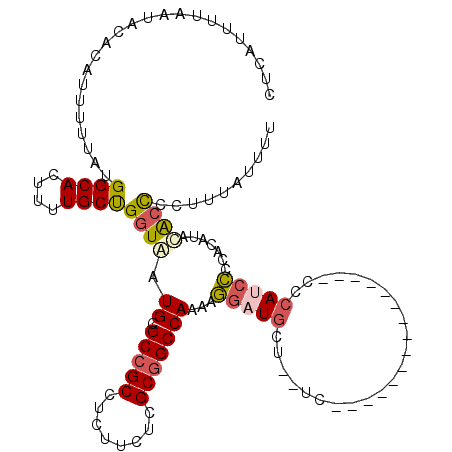
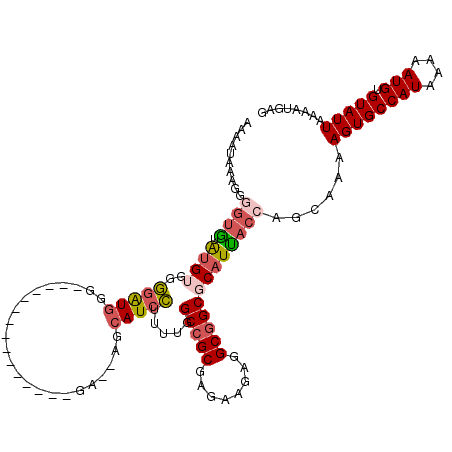
| Location | 12,802,714 – 12,802,817 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -16.26 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12802714 103 - 22407834 AAAAUAAAGGAGUGUAUGUGGGGAUGGG---------------GA--AGCAUCCUUUUGGCCGCGAGAAGAGGCGGCGCAUUACCAGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG ........(((((((....(((((((..---------------..--..)))))))...(((((........)))))))))).))......((((((((....))).)))))........ ( -27.40) >DroPse_CAF1 96182 114 - 1 ------GAGGGGCGAGGGGGGCGGUGGGCAAAGCCAGGAAUAAAAUGCGAAUGGGUUUGGCAGCGAUGAGAGGCGGCACAUUACCAGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG ------..((.((...(..((..(((.((...(((....((....(((.((.....)).)))...))....))).)).)))..))..)....)).))....................... ( -25.30) >DroSec_CAF1 68345 103 - 1 AAAAUAAAGGGGUAUAUGUGAGGAUGGG---------------GU--AGCAUCCUUUUGGCCGCGAGAAGAGGCGGCGCAUUACCAGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG ..........((((.(((((((((((..---------------..--..)))))))...(((((........)))))))))))))......((((((((....))).)))))........ ( -29.10) >DroSim_CAF1 70522 103 - 1 AAAAUAAAGGGGUAUAUGUGGGGAUGGG---------------GU--AGCAUCCUUUUGGCCGCGAGAAGAGGCGGCGCAUUACCAGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG ..........((((.(((((((((((..---------------..--..)))))))...(((((........)))))))))))))......((((((((....))).)))))........ ( -28.30) >DroYak_CAF1 71296 106 - 1 A----AAAGGAGUGUGUGGGUGGGUGGGUUU--------AUGAGA--GGCAUCCUUUUGGCCGCGAGAAGAGGCGGCGCAUCAGCGGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG .----.....((((..(..((.((((.....--------..((((--((...)))))).(((((........))))).)))).))((((....))))........)..))))........ ( -25.50) >consensus AAAAUAAAGGGGUGUAUGUGGGGAUGGG_______________GA__AGCAUCCUUUUGGCCGCGAGAAGAGGCGGCGCAUUACCAGCAAAAGUGCCAUAAAAAUGUGUAUUAAAAUGAG ..........((((.((((..(((((.......................))))).....(((((........)))))))))))))......((((((((....))).)))))........ (-16.26 = -17.06 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:11 2006