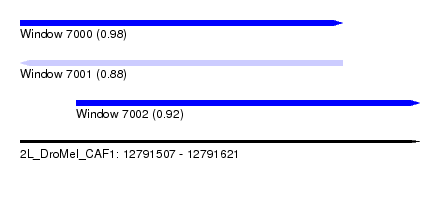
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,791,507 – 12,791,621 |
| Length | 114 |
| Max. P | 0.976877 |

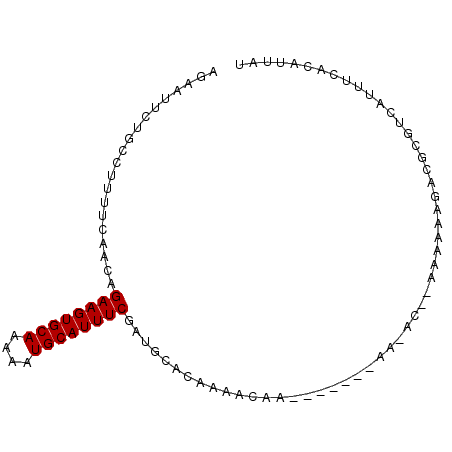
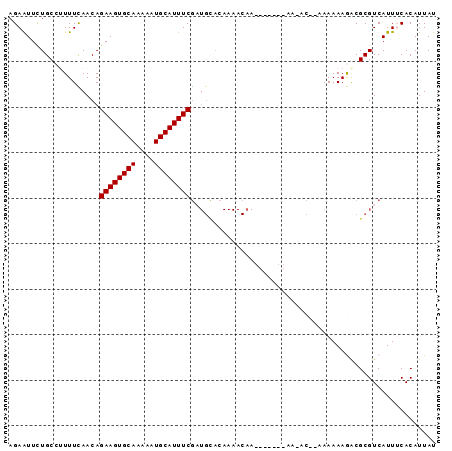
| Location | 12,791,507 – 12,791,599 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -15.08 |
| Consensus MFE | -9.50 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12791507 92 + 22407834 AGAAUUUUGCCUUUUCAACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACUAAAAAAGAAAAAC--AAAAAAGACGCGUCAUUUCACAUUAU .(((................((((((((....))))))))(((((.....................--.........)))))..)))....... ( -14.81) >DroSec_CAF1 60988 85 + 1 AGAAUUUUGCCUUUUCAACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACAA-------AAAGC--AAAAAAGGCGCGUCAUUUCACAUUAU .((....((((((((.....((((((((....))))))))..(((.........-------...))--).))))))))..))............ ( -19.60) >DroSim_CAF1 63127 70 + 1 AGAAUUCUGCCUUUUCAACAGAAGUGCAAAAAUGCAUUUCGU----------------------AC--AAAAAAGACGCGUCAUUUCACAUUAU .(((...((((((((.....((((((((....))))))))..----------------------..--..)))))..)))....)))....... ( -13.50) >DroEre_CAF1 62972 86 + 1 AGAGUUCUGCAAUUUUAACAGAAGUGCAA-AUUGCAUUUCGUUGCACAAAACAA-------AACACAAAAAAAAAACGCGUCAUUGCACAUUAU ...(((.((((((.......(((((((..-...)))))))))))))...)))..-------................(((....)))....... ( -15.01) >DroYak_CAF1 63583 82 + 1 AGAGUUCUGCCAUUUUAACAGAAGUGCAA-AAUGCAUUUCAAUGUACAAAACAA-----------CAAAAAAAAAACGCGUCAUUGCACAUUAU ....(((((.........)))))((((((-.((((.(((...(((........)-----------))......))).))))..))))))..... ( -12.50) >consensus AGAAUUCUGCCUUUUCAACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACAA_______AA_AC__AAAAAAGACGCGUCAUUUCACAUUAU ....................((((((((....))))))))...................................................... ( -9.50 = -9.50 + 0.00)

| Location | 12,791,507 – 12,791,599 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.54 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12791507 92 - 22407834 AUAAUGUGAAAUGACGCGUCUUUUUU--GUUUUUCUUUUUUAGUUUUGUGCAUCGAAAUGCAUUUUUGCACUUCUGUUGAAAAGGCAAAAUUCU ...(((((......)))))...((((--((((((..((..(((....(((((..((......))..)))))..)))..)))))))))))).... ( -15.30) >DroSec_CAF1 60988 85 - 1 AUAAUGUGAAAUGACGCGCCUUUUUU--GCUUU-------UUGUUUUGUGCAUCGAAAUGCAUUUUUGCACUUCUGUUGAAAAGGCAAAAUUCU .....(((......)))((((((((.--((...-------.(((...((((((....))))))....))).....)).))))))))........ ( -20.60) >DroSim_CAF1 63127 70 - 1 AUAAUGUGAAAUGACGCGUCUUUUUU--GU----------------------ACGAAAUGCAUUUUUGCACUUCUGUUGAAAAGGCAGAAUUCU .....(((......)))((((((((.--..----------------------(((((.((((....)))).))).)).))))))))........ ( -16.20) >DroEre_CAF1 62972 86 - 1 AUAAUGUGCAAUGACGCGUUUUUUUUUUGUGUU-------UUGUUUUGUGCAACGAAAUGCAAU-UUGCACUUCUGUUAAAAUUGCAGAACUCU .....((((((....(((((((....(((..(.-------.......)..))).)))))))...-))))))((((((.......)))))).... ( -20.50) >DroYak_CAF1 63583 82 - 1 AUAAUGUGCAAUGACGCGUUUUUUUUUUG-----------UUGUUUUGUACAUUGAAAUGCAUU-UUGCACUUCUGUUAAAAUGGCAGAACUCU .....((((((....(((((((.....((-----------(........)))..)))))))...-))))))(((((((.....))))))).... ( -19.80) >consensus AUAAUGUGAAAUGACGCGUCUUUUUU__GU_UU_______UUGUUUUGUGCAUCGAAAUGCAUUUUUGCACUUCUGUUGAAAAGGCAGAAUUCU .....(((......)))((((((((.................((.....))...(((.((((....)))).)))....))))))))........ (-11.48 = -11.54 + 0.06)

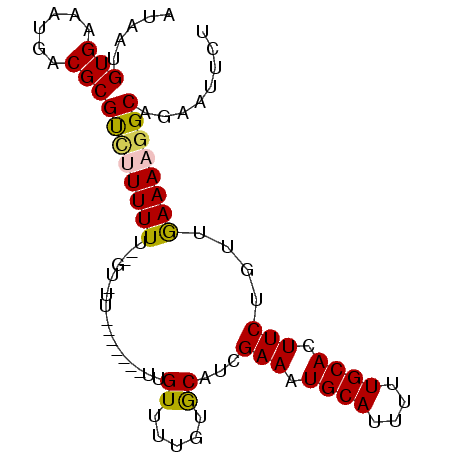
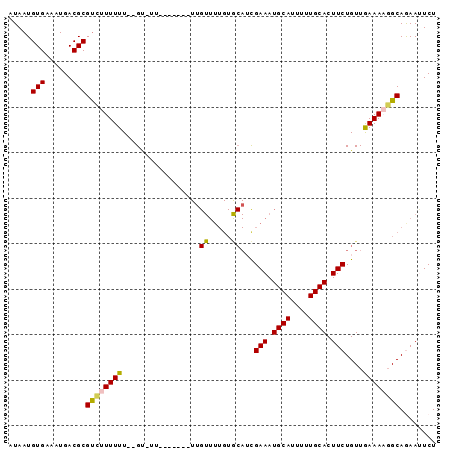
| Location | 12,791,523 – 12,791,621 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -16.20 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12791523 98 + 22407834 AACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACUAAAAAAGAAAAAC--AAAAAAGACGCGUCAUUUCACAUUAUAUCGCUGAUAGAGUAAUGCAUU ....((((((((....))))))))(((((.....................--.........)))))......((((((.((.......)))))))).... ( -17.11) >DroSec_CAF1 61004 91 + 1 AACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACAA-------AAAGC--AAAAAAGGCGCGUCAUUUCACAUUAUAUUGCUGAUAGAGUAAUGCAUU ....((((((((....))))))))(((((.(.......-------.....--.......).)))))...........(((((((.....))))))).... ( -18.71) >DroSim_CAF1 63143 76 + 1 AACAGAAGUGCAAAAAUGCAUUUCGU----------------------AC--AAAAAAGACGCGUCAUUUCACAUUAUAUCGCUGAUAGAGUAAUGCAUU ....((((((((....))))))))..----------------------..--.........((((.((((...((((......)))).)))).))))... ( -13.50) >DroEre_CAF1 62988 92 + 1 AACAGAAGUGCAA-AUUGCAUUUCGUUGCACAAAACAA-------AACACAAAAAAAAAACGCGUCAUUGCACAUUAUAUUGCUGAUAGAGUAAUGCAUU ....(((((((..-...)))))))..(((.........-------................(((....))).......((((((.....))))))))).. ( -17.20) >DroYak_CAF1 63599 88 + 1 AACAGAAGUGCAA-AAUGCAUUUCAAUGUACAAAACAA-----------CAAAAAAAAAACGCGUCAUUGCACAUUAUAUCGCUGAUAGAGUAAUGCAUU ....(((((((..-...)))))))((((((........-----------............(((....)))..........(((.....)))..)))))) ( -14.50) >consensus AACAGAAGUGCAAAAAUGCAUUUCGAUGCACAAAACAA_______AA_AC__AAAAAAGACGCGUCAUUUCACAUUAUAUCGCUGAUAGAGUAAUGCAUU ....((((((((....)))))))).....................................((((.((((...((((......)))).)))).))))... (-12.50 = -12.90 + 0.40)

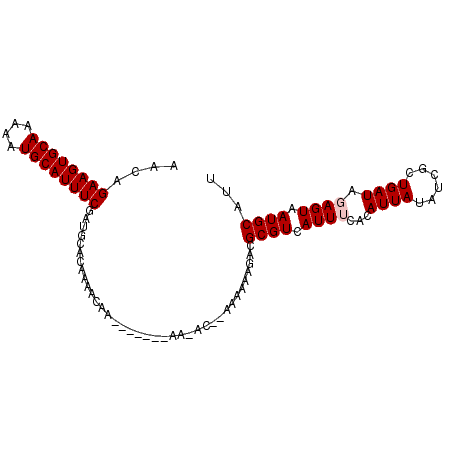
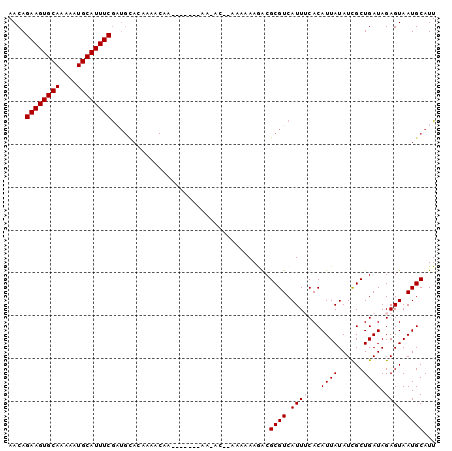
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:05 2006