
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,779,620 – 12,779,735 |
| Length | 115 |
| Max. P | 0.654828 |

| Location | 12,779,620 – 12,779,735 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.01 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
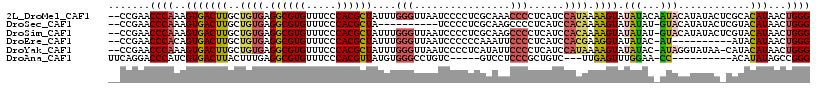
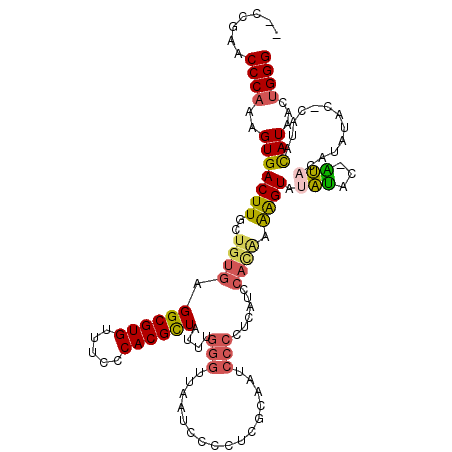
>2L_DroMel_CAF1 12779620 115 - 22407834 --CCGAACCCAAAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUAUUUGGGUUAAUCCCCUCGCAAACCCCUCAUCCAUAAAAGUAUAUACAAUACAUAUACUCGCACAUAACUGGG --.....((((..((....((((((((((((((.....))))).....(((.....))))))))).................(((((((.......))))))).)))....)))))) ( -29.20) >DroSec_CAF1 56523 104 - 1 --CCGAACCCAAAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUA----------UCCCCUCGCAAGCCCCUCAUCCACAAAAGUAUAUAU-GUACAUAUACUCGUACAUAACUGGG --.....((((..((((...(((((((((((((.....)))))..----------....))))).)))...)))).............(((-((((........)))))))..)))) ( -28.10) >DroSim_CAF1 57160 114 - 1 --CCGAACCCAAAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUAUUUGGGUUAAUCCCCUCGCAAGCCCCUCAUCCACAAAAGUAUAUAU-GUACAUAUACUCGUACAUAACUGGG --.....((((..((((...(((((((((((((.....))))).....(((.....)))))))).)))...)))).............(((-((((........)))))))..)))) ( -34.00) >DroEre_CAF1 58473 104 - 1 --CCGAACCCACAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUAUUUGGGUUAAUCCCCCCAAAUUCCCCUCAUCCACGAAGGUAUAUAC-AU----------AUACAUAACUGGG --.......((((((.....)))))).((((((.....))))))(((((((........))))))).........(((.....(((((...-.)----------)))).....))). ( -27.70) >DroYak_CAF1 58974 113 - 1 --CCGAACCCAAAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUAUUUGGGUUAAUCCCCUCAUAUUCCCCUCAUCCAUAAAAGUAUAUAC-AUAGGUAUAA-CAUACAUAACUGGG --.....((((..(((((((...((((((((((.....)))))(((..(((......)))..))).....)))))......)))).(((((-....))))).-....)))...)))) ( -25.20) >DroAna_CAF1 55952 98 - 1 UUCAGGACCCAUCGUGACUUACUUUGAGGCGUGUUUCCCACGUUAUGUGGGCCUGUC-----GUCCUCCCGCUGUC---UUGAGUUUGGAA-CC----------ACAUAUAGCCGGG ..((((.((((.((((.(((.....)))(((((.....)))))))))))))))))..-----.....((((((((.---.((.((.....)-))----------)...)))).)))) ( -27.30) >consensus __CCGAACCCAAAGUGACUUGCUGUGAGGCGUGUUUCCCACGCUAUUUGGGUUAAUCCCCUCGCAAUCCCCUCAUCCACAAAAGUAUAUAC_AUACAUAUAC_CAUACAUAACUGGG .......((((..(((((((..((((.((((((.....))))))....(((................)))......)))).)))).(((...)))............)))...)))) (-13.84 = -13.96 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:00 2006