| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,774,357 – 12,774,451 |
| Length | 94 |
| Max. P | 0.560693 |

| Location | 12,774,357 – 12,774,451 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12774357 94 - 22407834 UCUGAGGGUGACC-C-UCGAG--------GCACCGCUGGCAAACCAAUAGAA-AAAUGUUUGUUUGUGGCAGC-AUC-------AGGA----GCUGGAGCAGCAGUUGGUGAUGUUG ((((((((...))-)-)).))--------)..((((..((((((........-....))))))..))))((((-(((-------(..(----((((......)))))..)))))))) ( -32.90) >DroPse_CAF1 70762 100 - 1 UCUGAGGGUGACCCC-UCGGG--------UUGCUGCUGGCAAACCAAUAGAAAAAAUGUUUGUUUGCGGCUGCCGUC-------AGGAGCA-GCAGGAGCAGCAGGAGCAGCAGUUG .(((((((....)))-))))(--------((.((((((((((((.((((.......)))).)))))).((((((...-------..).)))-)).....)))))).)))........ ( -42.70) >DroEre_CAF1 53426 94 - 1 UCUGAGGGUGACC-C-UCGGG--------GCUCCGCUGGCAAACCAAUAGAA-AAAUGUUUGUUUGUGGCAGC-AUC-------GGGA----GCUGGAGCAGCAGUUGGUGAUGUUG ((((((((...))-)-)))))--------...((((..((((((........-....))))))..))))((((-(((-------(..(----((((......)))))..)))))))) ( -35.20) >DroYak_CAF1 53749 94 - 1 UCUGAGGGUGACC-C-UCGGG--------GCUCCGCUGGCAAACCAAUAGAA-AAAUGUUUGUUUGUGGCAGC-AUC-------AGGA----GCCGGAGCAGCAGUUGCUGAUGUUG .(((((((...))-)-))))(--------(((((((((((((((.((((...-...)))).))))))..))))-...-------.)))----)))....((((....))))...... ( -38.80) >DroAna_CAF1 51200 114 - 1 UCUGAGGGUGACC-CCUCCGGGAGUCUCUGCUCUGCUGGCAAACCAAUAGAA-AAAUGUUUGUUUGCGGCAGC-AUCAGGCAUCAGGAGCAAGUCGGAGCAGCAGUUGGUGCCAUUG (((((((....))-.((((..((((((.((((((((..((((((........-....))))))..)))).)))-)..)))).)).))))....)))))(((.(....).)))..... ( -39.40) >DroPer_CAF1 71955 100 - 1 UCUGAGGGUGACCCC-UCGGG--------GUGCUGCUGGCAAACCAAUAGAAAAAAUGUUUGUUUGCGGCUGCCGUC-------AGGAGCA-GCAGGAGCAGCAGGAGCAGCAGUUG .(((((((....)))-))))(--------.(.((((((((((((.((((.......)))).)))))).((((((...-------..).)))-)).....)))))).).)........ ( -40.60) >consensus UCUGAGGGUGACC_C_UCGGG________GCUCCGCUGGCAAACCAAUAGAA_AAAUGUUUGUUUGCGGCAGC_AUC_______AGGA____GCAGGAGCAGCAGUUGCUGAUGUUG .((((((....))...))))..............((((((((((.((((.......)))).))))))..((((...................))))...)))).............. (-20.00 = -20.06 + 0.06)


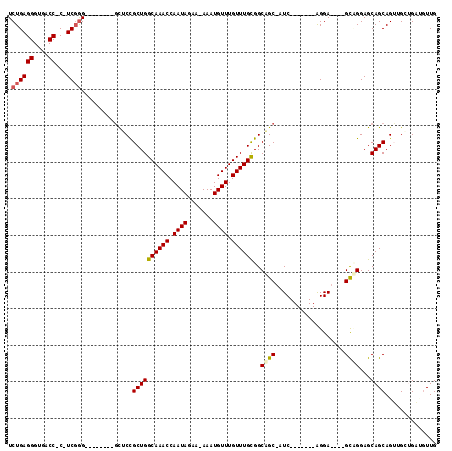
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:58 2006