| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,769,293 – 12,769,385 |
| Length | 92 |
| Max. P | 0.856467 |

| Location | 12,769,293 – 12,769,385 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12769293 92 + 22407834 ACGUUCAUCA--UCGCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA--GCAGGUGGG------------GAC ..((((.(((--((((((((((.((((((((((....)))((((((((((....)).))))))))..))))))).))).)))--)).))))))------------))) ( -37.10) >DroSec_CAF1 46303 100 + 1 ACGUUCAUCA--UCGCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA--GCAGGGGGGUG----GGGGCAGGC ..((((.(((--((.((..(((((.(((..(...(((.(.((((((((((....)).)))))))).).))))..)))..)))--))..)).))))----))))).... ( -39.80) >DroSim_CAF1 46548 99 + 1 ACGUUCAUCA--UCGCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA--GCAGGGGGAUG----UGGGU-GGC .((..((.((--((.((..(((((.(((..(...(((.(.((((((((((....)).)))))))).).))))..)))..)))--))..)).))))----))..)-).. ( -40.90) >DroEre_CAF1 48243 80 + 1 --------------GCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA--GCGGGCGCG------------GGC --------------((((((((((.(((..(...(((.(.((((((((((....)).)))))))).).))))..)))..)))--))))).)).------------... ( -32.10) >DroYak_CAF1 48485 89 + 1 ACGUUCAUCA--UCGCUUCGUUUCGCUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA--GCGGG---G------------GGC ..........--.(.(((((((((((((..(...(((.(.((((((((((....)).)))))))).).))))..)))).)))--)))))---)------------.). ( -37.10) >DroAna_CAF1 46273 107 + 1 AUUUUCAUCAGCUUGCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUCGAAAUGUGUGCUGCAGACGCAGUGCAGCAGCAUGGUAGGGGUUUGCCAAUGGGU-UGG ...(((((..((..(((((...((((((((((..(((.(.((((((((.(.....).)))))))).).))).)))).)))..)))..)))))..))..))))).-... ( -36.30) >consensus ACGUUCAUCA__UCGCUUCGUUUCACUGUGCAAAUGUUUGUGCAGCACUAGAAAUGUGUGUUGCAGACGCAGUGCAGCAGGA__GCAGGGGGG____________GGC ...............(((.((....(((((((..(((.(.((((((((.........)))))))).).))).)))).)))....)))))................... (-23.55 = -23.05 + -0.50)


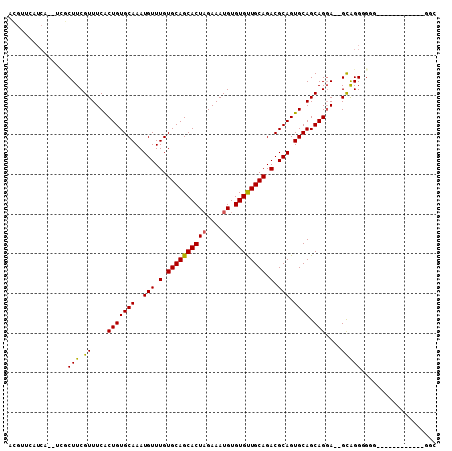
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:55 2006