
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
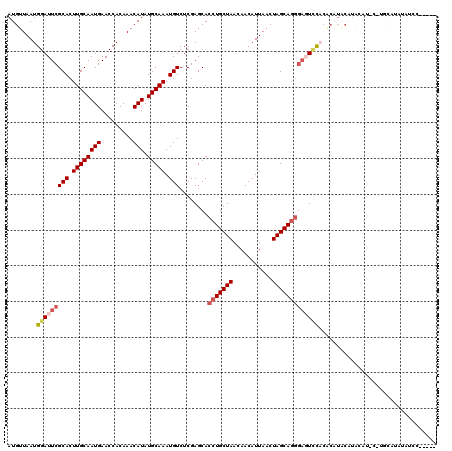
| Location | 12,769,055 – 12,769,166 |
| Length | 111 |
| Max. P | 0.940475 |

| Location | 12,769,055 – 12,769,166 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -18.07 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
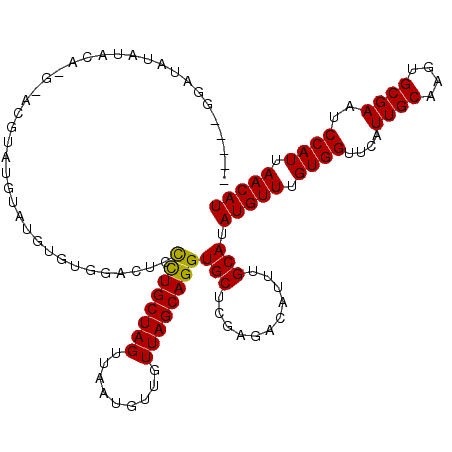
>2L_DroMel_CAF1 12769055 111 + 22407834 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGGCAGUCCACACAUACAUACGU----AUGUAUAUCC----- .......((((((.(((.((((((((.........))).))))).)))........(((((((...........))))))).)))))).((((((....))----))))......----- ( -26.10) >DroPse_CAF1 60903 113 + 1 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGAGUCUCUC--CCUCCUCACCCCCCUCUCCACCUCC----- .......((((...(((.((((((((.........))).))))).)))...(((.((((((((...........)))))).)))))..--................)))).....----- ( -21.20) >DroSec_CAF1 46061 115 + 1 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGGGAGUACACACAUGCAUACGUGUGUAUGUAUAUCC----- .((((..(((.(((((....))...))))))..))))(((((..............(((((((...........)))))))..((((((((........)))))))))))))...----- ( -29.90) >DroEre_CAF1 48014 108 + 1 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGGGAGUCCACACAUACGUGUAUCCUGGCA------------ .(((((..(((((((((.((((((((.........))).))))).)))........(((((((...........)))))))))))))((((....))))....)))))------------ ( -29.80) >DroYak_CAF1 47224 120 + 1 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGGGAGUCUACGCAUACAUACAUACAUGCAUAUAGCCAUCCC ..((((.((((((((((.((((((((.........))).))))).)))........(((((((...........)))))))))))))).((((..........))))...))))...... ( -27.20) >DroAna_CAF1 46054 97 + 1 AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAAGGACUUCCCCA--------------GCA----CCC----- ..(((((((((...(((.((((((((.........))).))))).))).)).(((....))).....)))))))..((..((.....))..--------------)).----...----- ( -18.90) >consensus AUGUUAAUGGAUUCGCACUUGCAAUGAACCACAAACAUAUGCAAAUGUCUCGAGCACCUGCUAACAACAUUAACUAGCAGGGAGUCCACACAUACAUACAU_C_UGCAUAUAUCC_____ ........(((((((((.((((((((.........))).))))).)))........(((((((...........)))))))))))))................................. (-18.07 = -18.90 + 0.83)


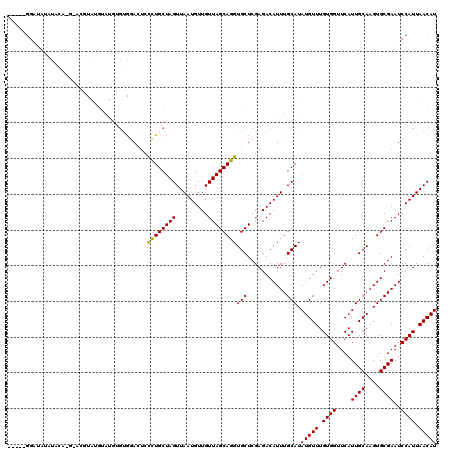
| Location | 12,769,055 – 12,769,166 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12769055 111 - 22407834 -----GGAUAUACAU----ACGUAUGUAUGUGUGGACUGCCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU -----((((.((((.----((((((((((((.(.((..(((((((((.........)))))))))..)).).))))..)))))))).)))).....((((....))))))))........ ( -38.60) >DroPse_CAF1 60903 113 - 1 -----GGAGGUGGAGAGGGGGGUGAGGAGG--GAGAGACUCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU -----(..((((((................--..(((((.(((((((.........))))))))).)))..(.((((((((.(((.........))))))))))))...))))))..).. ( -29.10) >DroSec_CAF1 46061 115 - 1 -----GGAUAUACAUACACACGUAUGCAUGUGUGUACUCCCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU -----((((.....((((.(((((((((...((((.(((((((((((.........)))))))).....))))))).))))))))).)))).....((((....))))))))........ ( -37.50) >DroEre_CAF1 48014 108 - 1 ------------UGCCAGGAUACACGUAUGUGUGGACUCCCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU ------------.....(((((((..((((((..(((((((((((((.........)))))))).....)))...))..))))))..)))......((((....))))))))........ ( -28.70) >DroYak_CAF1 47224 120 - 1 GGGAUGGCUAUAUGCAUGUAUGUAUGUAUGCGUAGACUCCCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU .(((((((((((.((((((..(.((((...((.((....((((((((.........))))))))..))))..)))).)..)))))).)))))))..((((....))))))))........ ( -33.80) >DroAna_CAF1 46054 97 - 1 -----GGG----UGC--------------UGGGGAAGUCCUUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU -----(((----(((--------------(.(((....(((.(((((.........))))))))..))).)).)))))....(((((.((((....((((....))))..)))).))))) ( -25.30) >consensus _____GGAUAUAUACA_G_ACGUAUGUAUGUGUGGACUCCCUGCUAGUUAAUGUUGUUAGCAGGUGCUCGAGACAUUUGCAUAUGUUUGUGGUUCAUUGCAAGUGCGAAUCCAUUAACAU .......................................((((((((.........))))))))(((...........))).(((((.((((....((((....))))..)))).))))) (-20.79 = -20.52 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:54 2006