| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,767,155 – 12,767,251 |
| Length | 96 |
| Max. P | 0.854733 |

| Location | 12,767,155 – 12,767,251 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
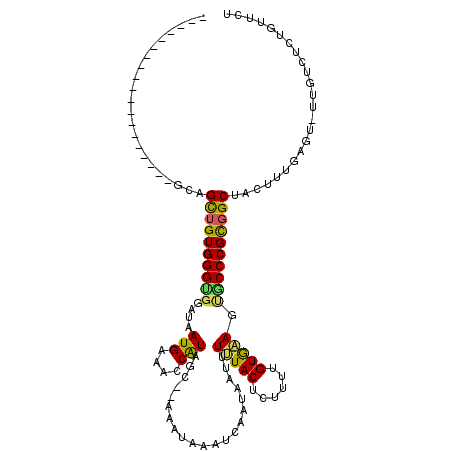
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
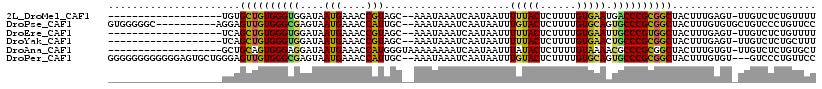
>2L_DroMel_CAF1 12767155 96 + 22407834 -------------------UGUGCUGUGGGUGGAUAAUGAAACCGUAGC--AAAUAAAUCAAUAAUUUUUACUCUUUUGUGAAUGACCCGCGGCUACUUUGAGU-UUGUCUCUGUUUU -------------------.(((((((((((.(...(..(((..((((.--((((.........))))))))..)))..)...).))))))))).))...(((.-....)))...... ( -22.10) >DroPse_CAF1 59415 106 + 1 GUGGGGGC----------AGGAGUUGUGGGCGAGUAAUGAAACCAUUGC--AAAUAAAUCAAUAAUUUGUACUCUUUUGUGCAGUGCCCGCGGCUACUUUGUGUGCUGUCCCUGUUCC (..(((((----------((.(((((((((((.((((((....))))))--...............((((((......)))))))))))))))))((.....)).)))))))..)... ( -42.40) >DroEre_CAF1 46127 96 + 1 -------------------UCAGCUGUGGGUGGAUAAUGAAACCGUAGC--AAAUAAAUCAAUAAUUUUUACUCUUUUGUGAAUUGCCCGUGGCUACUUUGAGU-UUGUCUCUGUUUU -------------------..(((..((((..(...(..(((..((((.--((((.........))))))))..)))..)...)..))))..))).....(((.-....)))...... ( -21.40) >DroYak_CAF1 45312 96 + 1 -------------------UCAGCUGUGGGUGGAUAAUGAAACCGUAGC--AAAUAAAUCAAUAAUUUUUACUCUUUUGUGAACUGCCCGCGGCUACUUUGAGU-UUGUCUCUGCUUU -------------------..(((((((((..(...(..(((..((((.--((((.........))))))))..)))..)...)..))))))))).....(((.-....)))...... ( -25.50) >DroAna_CAF1 44062 98 + 1 -------------------GCUGCAGUGGGAGGAUAAUGAAACCAUGGGUAAAAAAAAUCAAUAAUUUAUACUCUUUUGUAAAACGCCCGCGGCUACUUUGUGU-UUGUCUCUGUGCU -------------------((.((((.(((((.((((.....((.(((((.....((((.....)))).(((......)))....))))).)).....)))).)-)).)).)))))). ( -17.80) >DroPer_CAF1 59113 113 + 1 GGGGGGGGGGGGAGUGCUGGGAGUUGUGGGCGAGUAAUGAAACCAUUGC--AAAUAAAUCAAUAAUUUGUACUCUUUUGUGCAGUGCCCGCGGCUACUUUGUGU---GUCCCUGUUCC ..((((.((((..(..(..(((((((((((((.((((((....))))))--...............((((((......))))))))))))))))).))..)..)---..)))).)))) ( -46.70) >consensus ___________________GCAGCUGUGGGUGGAUAAUGAAACCAUAGC__AAAUAAAUCAAUAAUUUUUACUCUUUUGUGAAGUGCCCGCGGCUACUUUGAGU_UUGUCUCUGUUCU ......................((((((((((....(((....))).....................(((((......))))).))))))))))........................ (-17.90 = -17.10 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:50 2006