| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,765,204 – 12,765,300 |
| Length | 96 |
| Max. P | 0.817980 |

| Location | 12,765,204 – 12,765,300 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
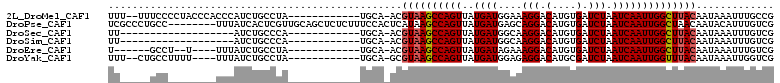
>2L_DroMel_CAF1 12765204 96 - 22407834 UUU--UUUCCCCUACCCACCCAUCUGCCUA------------UGCA-ACGUAAGCCAGUUAUGAUGGAAAGGACAUGUGAUCUAAUCAAUUGGCUUACAAUAAAUUUGCCG ...--...................(((...------------.)))-..((((((((((..(((((((............))).))))))))))))))............. ( -17.50) >DroPse_CAF1 56438 103 - 1 UCGCCCUGCC--------UUUAUCACUCGUUGCAGCUCUCUUUCCACUCAUAAGCCAGUUAUGAUGAGCAGGACAUGUGAUCUAAUCAAUUGGCUAACAAUACAUUUGUCG ..(((.((..--------...(((((..(((...((((.........((((((.....)))))).))))..)))..))))).....))...)))..((((.....)))).. ( -16.60) >DroSec_CAF1 42225 79 - 1 UU-------------------AUCUGCCCA------------UGCA-ACGUAAGCCAGUUAUGAUGGCAAGGACAUGUGAUCUAAUCAAUUGGCUUACAAUAAAUUUGUCG ..-------------------...(((...------------.)))-..(((((((((((..(((.(((......))).))).....)))))))))))............. ( -21.00) >DroSim_CAF1 42492 79 - 1 UU-------------------AUCUGCCCA------------UGCA-ACGUAAGCCAGUUAUGAUGGCAAGGACAUGUGAUCUAAUCAAUUGGCUUACAAUAAAUUUGUCG ..-------------------...(((...------------.)))-..(((((((((((..(((.(((......))).))).....)))))))))))............. ( -21.00) >DroEre_CAF1 42966 86 - 1 U------GCCU--U----UUUAUCUGCCUA------------UGCA-ACGUAAGCCAGUUAUGAUAGAAAGGACAUGUGAUCUAAUCAAUUGGCUUACAAUAAAUUUGUCG .------....--.----......(((...------------.)))-..((((((((((..(((((((............))).))))))))))))))............. ( -17.80) >DroYak_CAF1 43323 92 - 1 UUU--CUGCCUUUU----UUUAUCUGCCUA------------UGCA-GCGUAAGCCAGUUAUGAUGGAGAGGACAUGCGAUCUAAUCAAUUGGUUUACAAUAAAUUGGUCG ...--..(((....----(((((((((...------------.)))-).((((((((((..(((((((..(......)..))).)))))))))))))).)))))..))).. ( -22.70) >consensus UU_____GCC________UUUAUCUGCCUA____________UGCA_ACGUAAGCCAGUUAUGAUGGAAAGGACAUGUGAUCUAAUCAAUUGGCUUACAAUAAAUUUGUCG .................................................((((((((((..((((....(((.(....).))).))))))))))))))............. (-14.40 = -14.60 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:49 2006