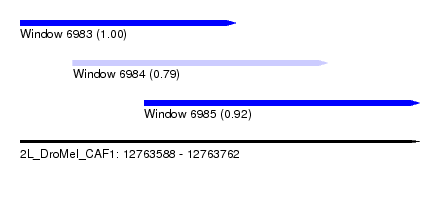
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,763,588 – 12,763,762 |
| Length | 174 |
| Max. P | 0.999371 |

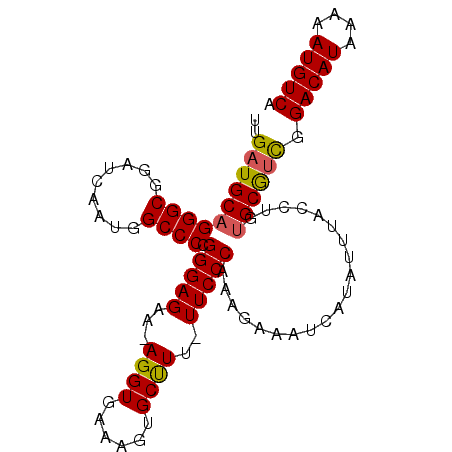
| Location | 12,763,588 – 12,763,682 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12763588 94 + 22407834 UUGAUGCCGGGCGGAUCAAUGGCCCCGGGAGAA-AGGUGAAAGUGCUU--UUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCA .((((((((((..(((..((((...(.((.(((-((((......))))--)))))...)...)))))))..)))).))))))(((((....))))). ( -29.70) >DroSec_CAF1 40744 96 + 1 UUGAUGCAGGGCGGAUCAAUGGCCCCGGGAGAA-AGGUGAAAGUGCUUUUUUCCCAAAGAAAUCAUAUUUAUCUGUGCGGCGGACAUAAAAAUGUCA ....(((.(.((((((.((((.....(((((((-((((......)))))))))))...(....).)))).)))))).).)))(((((....))))). ( -30.80) >DroSim_CAF1 40996 96 + 1 UUGAUGCAGGGCGGAUCAAUGGCCCCGGGAGAA-AGGUGAAAGUGCUUUUUUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCA .(((((((((((.........)))).(((((((-((((......)))))))))))....................)))))))(((((....))))). ( -34.30) >DroEre_CAF1 41565 95 + 1 UUGAUGCAGGGCGGAUCAAUGGCCCCGGGAGAA-AGGUUAAAGUGCUUU-UUCCCAAAGAAAUCAUAUUUGCCUGUGCAUUCGACAUAAAAAUGUCA ..((((((((((((((....(....)(((((((-((.........))))-)))))...........)))))))).)))))).(((((....))))). ( -30.40) >DroYak_CAF1 41851 96 + 1 UUGAUGCAGGGCGGAUCAAUGGCCCCGGGAGAAAAGGUGAAAGUGCCUU-UUCCCAAAGAAAUCAUAUUUACCUGUGCGUUGGACAUAAAAAUGUCA ..((((((((((.........))))..((.((((((((......)))))-)))))....................)))))).(((((....))))). ( -29.30) >consensus UUGAUGCAGGGCGGAUCAAUGGCCCCGGGAGAA_AGGUGAAAGUGCUUU_UUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCA ..((((((((((.........)))).(((((...((((......))))..)))))....................)))))).(((((....))))). (-26.70 = -26.54 + -0.16)

| Location | 12,763,611 – 12,763,722 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12763611 111 + 22407834 CCCGGGAGAA-AGGUGAAAGUGCUU--UUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAGGAAAAAAUGGCAGGGCGAGGAAGGAGGAAUCAGGUGUAU ....((.(((-((((......))))--)))))..............((((((..(.((.(((((....)))))...........((...)).......)).)...))))))... ( -24.00) >DroSec_CAF1 40767 112 + 1 CCCGGGAGAA-AGGUGAAAGUGCUUUUUUCCCAAAGAAAUCAUAUUUAUCUGUGCGGCGGACAUAAAAAUGUCAG-AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAU ...(((((((-((((......)))))))))))...........(((((((((..(.((...........(((((.-......)))))..........))..)...))))))))) ( -29.10) >DroSim_CAF1 41019 112 + 1 CCCGGGAGAA-AGGUGAAAGUGCUUUUUUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAG-AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAU ...(((((((-((((......)))))))))))...........(((((((((....((.(((((....)))))..-.............((......))..))..))))))))) ( -32.40) >DroEre_CAF1 41588 109 + 1 CCCGGGAGAA-AGGUUAAAGUGCUUU-UUCCCAAAGAAAUCAUAUUUGCCUGUGCAUUCGACAUAAAAAUGUCAG--GAAAAU-GCAGGGCGAGUAAGCGGCAAUCAGGUGGAU .(((((((((-((.........))))-)))))..........(((((((((.((((((.(((((....)))))..--...)))-))))))))))))..............)).. ( -33.40) >DroYak_CAF1 41874 111 + 1 CCCGGGAGAAAAGGUGAAAGUGCCUU-UUCCCAAAGAAAUCAUAUUUACCUGUGCGUUGGACAUAAAAAUGUCAG-GGAAAAU-GCAGGGCGAGGAAACGGAAAUCAGGUGGAU ....((.((((((((......)))))-)))))...........(((((((((((((((.(((((....)))))..-....)))-)))......(....)......))))))))) ( -33.10) >consensus CCCGGGAGAA_AGGUGAAAGUGCUUU_UUCCCAAAGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAG_AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAU ...(((((...((((......))))..)))))...........(((((((((.......(((((....)))))...........((...))..............))))))))) (-22.40 = -22.12 + -0.28)

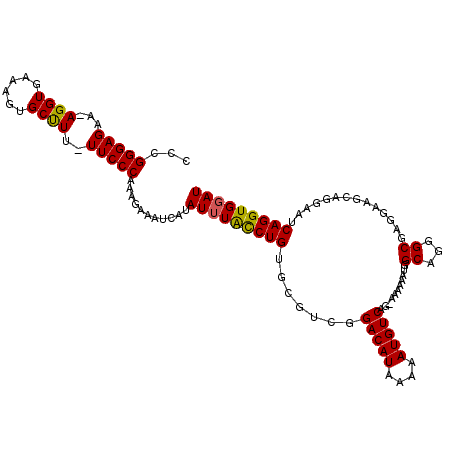
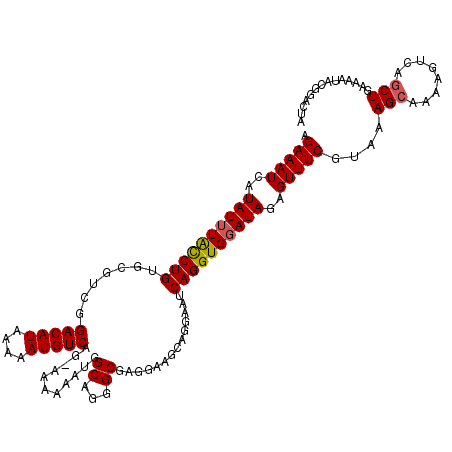
| Location | 12,763,642 – 12,763,762 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -20.84 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12763642 120 + 22407834 AGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAGGAAAAAAUGGCAGGGCGAGGAAGGAGGAAUCAGGUGUAUAGAGUUUCGUAAAGCAAAAGUCAGCUGAAAAUACGGACUA .(((((..(((.((((((..(.((.(((((....)))))...........((...)).......)).)...)))))).)))..)))))(((.(((........))).....)))...... ( -24.20) >DroSec_CAF1 40800 119 + 1 AGAAAUCAUAUUUAUCUGUGCGGCGGACAUAAAAAUGUCAG-AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAUAGAGUUUCGUAAAGCAAAAGUCAGCUGAAAAUACGGACUA ..............((((((((((.(((.......(((((.-......)))))..((.(.(((((....(((.....)))...))))).)...))....))).))))....))))))... ( -24.90) >DroSim_CAF1 41052 119 + 1 AGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAG-AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAUAGAGUUUCGUAAAGCAAAAGUCAGCUGAAAAUACGGACUA .(((((..((((((((((....((.(((((....)))))..-.............((......))..))..))))))))))..)))))(((.(((........))).....)))...... ( -27.80) >DroEre_CAF1 41620 117 + 1 AGAAAUCAUAUUUGCCUGUGCAUUCGACAUAAAAAUGUCAG--GAAAAU-GCAGGGCGAGUAAGCGGCAAUCAGGUGGAUAGGGUUUCAUAAAGGAAGAGUCAGCUGACAAGACAAAAUA .((((((.((((..((((((((((.(((((....)))))..--...)))-))).(.((......)).)...))))..)))).))))))...........(((....)))........... ( -32.00) >DroYak_CAF1 41907 117 + 1 AGAAAUCAUAUUUACCUGUGCGUUGGACAUAAAAAUGUCAG-GGAAAAU-GCAGGGCGAGGAAACGGAAAUCAGGUGGAUAGAGUUUCAUAAAGCAAGACUCAGCUGAAAAGG-AGUAUA .(((((..((((((((((((((((.(((((....)))))..-....)))-)))......(....)......))))))))))..)))))..........((((..((....)))-)))... ( -32.20) >consensus AGAAAUCAUAUUUACCUGUGCGUCGGACAUAAAAAUGUCAG_AAAAAAUGGCAGGGCGAGGAAGCAGGAAUCAGGUGGAUAGAGUUUCGUAAAGCAAAAGUCAGCUGAAAAUACGGACUA .(((((..((((((((((.......(((((....)))))...........((...))..............))))))))))..)))))....(((........))).............. (-20.84 = -20.92 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:48 2006