
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,759,823 – 12,759,933 |
| Length | 110 |
| Max. P | 0.617222 |

| Location | 12,759,823 – 12,759,933 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -17.51 |
| Consensus MFE | -11.88 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12759823 110 + 22407834 AUUUUAAUUAAUCACUGGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAUCCCUUCGGUCU-CC---UCUC-----CAAUCAGCCUCUA-AAAAUCCC .............(((((((((((.....)))..(((((((((.........))).))))))...........))))))))..-..---....-----.............-........ ( -17.30) >DroPse_CAF1 49339 112 + 1 AUUUUAAUUAAUUACUAGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAAACUUCUGCCCAGCCCCCCCCC-----CCGAC---CCCCACCAGACCCC ...................((((.......(((..((((......))))..)))((......................))...))))......-----.....---.............. ( -12.45) >DroEre_CAF1 37806 110 + 1 AUUUUAAUUAAUCACUGGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAUCCCUUCGGUCU-CC---CCUC-----CAAUCAGCCUCUA-AAAAUCCU ...............(((((((........(((..((((......))))..)))....................(....)...-.)---))))-----))...........-........ ( -18.10) >DroYak_CAF1 38112 110 + 1 AUUUUAAUUAAUCACUGGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAUCCCUUUGGUCU-CC---CCUC-----CAAUCAGCCUCUA-AAAAUCCC ...............(((((((........(((..((((......))))..)))............(((........)))...-.)---))))-----))...........-........ ( -19.60) >DroAna_CAF1 35215 109 + 1 AUUUUAAUUAAUCACUGGAGGGCAUCAAUUGCAUUUUGAAUGCUAAUAAUUUGCGCAAUAAAUAUUCCAUAACAUUUUCGUUG-CCA--CCUCCGGGGCAAUC---CCCCA-ACAA---- ................((((((((...(((((((.....)))).)))....)))((((..((..............))..)))-)..--)))))((((.....---)))).-....---- ( -25.14) >DroPer_CAF1 49091 109 + 1 AUUUUAAUUAAUCACUAGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAAACUUCUGCCCAGCCCC---CC-----CAGAC---CCCCACCAGACCCC ...................((((.......(((..((((......))))..)))((......................))...)))).---..-----.....---.............. ( -12.45) >consensus AUUUUAAUUAAUCACUGGAGGGCAUCAAUUGCAUUUUGUAUGCUAAUAAUUUGCGCUAUAAAUAUUCCAUAACCCUUUGGUCU_CC___CCUC_____CAAUC___CCCCA_AAAAUCCC .............(((((((((((.....)))..(((((((((.........))).))))))...........))))))))....................................... (-11.88 = -12.30 + 0.42)

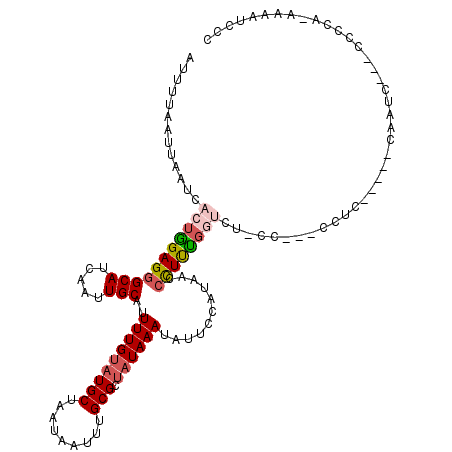
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:43 2006