
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
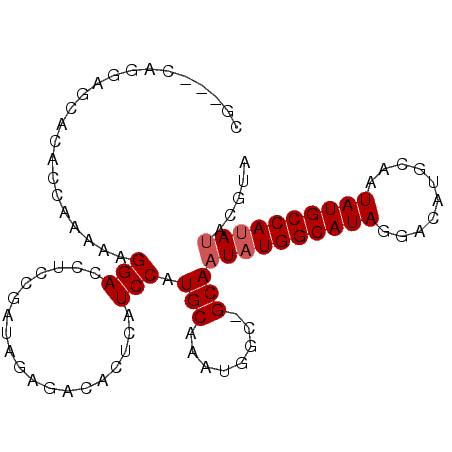
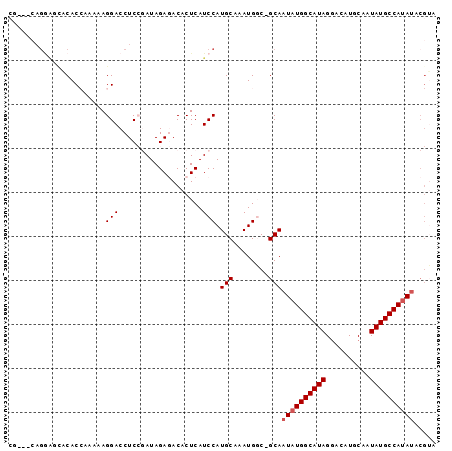
| Location | 12,754,544 – 12,754,635 |
| Length | 91 |
| Max. P | 0.847194 |

| Location | 12,754,544 – 12,754,635 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -9.13 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
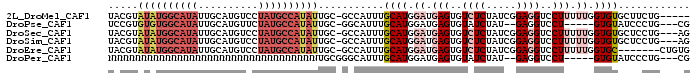
>2L_DroMel_CAF1 12754544 91 + 22407834 -----CAGAAGCACACCAAAAAGGACCUCCGAUAGAGACACUCAUCCAUGCAAAUGGC-GCAAUAUGGCAUAGGACAUGCAAUAUGCCAUAUACGUA -----.....(((.........((....))(((.(((...))))))..))).....((-(..((((((((((..........)))))))))).))). ( -20.50) >DroPse_CAF1 40279 86 + 1 CG---CAGGGAUACAC-----AGGACCUC--AUAGAUACACUCAUCCAUGCAAAUGCC-GCAAUAUGGCAUAGAACAUGCAAUAUGCCACACACGGA .(---((.((((....-----..(....)--...((.....)))))).))).....((-(.....(((((((..........)))))))....))). ( -17.60) >DroSec_CAF1 31590 93 + 1 CU---CAGGAGCACACCAAAAAGGACCUCCGAUAGAGACACUCAUCCAUGCAAAUGGC-GCAAUAUGGCAUAGGACAUGCAAUAUGCCAUAUACGUA ((---(.((((....((.....))..))))....)))((......((((....)))).-...((((((((((..........))))))))))..)). ( -22.30) >DroSim_CAF1 31833 93 + 1 CU---CAGGAGCACACCAAAAAGGACCUCCGAUAGAGACACUCAUCCAUGCAAAUGGC-GCAAUAUGGCAUAGGACAUGCAAUAUGCCAUAUACGUA ((---(.((((....((.....))..))))....)))((......((((....)))).-...((((((((((..........))))))))))..)). ( -22.30) >DroEre_CAF1 32577 89 + 1 CACAG-------GCACCAAAAAGGACCUCCGAUAGAGACACUCAUCCAUGCAAAUGGC-GCAAUAUGGCAUAGGACAUGCAAUAUGCCAUAUACGUA .((..-------((.(((....((....))(((.(((...))))))........))).-)).((((((((((..........))))))))))..)). ( -22.30) >DroPer_CAF1 39781 87 + 1 CG---CAGGGAUACAC-----AGGACCUC--AUAGAUACACUCAUCCAUGCAAAUGCCCGCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .(---(.(((.....(-----((((....--.............))).))......))))).................................... ( -7.73) >consensus CG___CAGGAGCACACCAAAAAGGACCUCCGAUAGAGACACUCAUCCAUGCAAAUGGC_GCAAUAUGGCAUAGGACAUGCAAUAUGCCAUAUACGUA ......................(((...................))).(((........)))((((((((((..........))))))))))..... ( -9.13 = -9.88 + 0.75)

| Location | 12,754,544 – 12,754,635 |
|---|---|
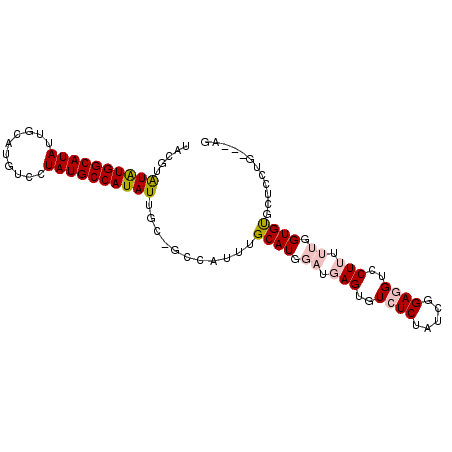
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -15.68 |
| Energy contribution | -17.07 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12754544 91 - 22407834 UACGUAUAUGGCAUAUUGCAUGUCCUAUGCCAUAUUGC-GCCAUUUGCAUGGAUGAGUGUCUCUAUCGGAGGUCCUUUUUGGUGUGCUUCUG----- ..(((((((((((((..........))))))))).)))-)......((((.((.(((..((((.....))))..))).)).)))).......----- ( -26.10) >DroPse_CAF1 40279 86 - 1 UCCGUGUGUGGCAUAUUGCAUGUUCUAUGCCAUAUUGC-GGCAUUUGCAUGGAUGAGUGUAUCUAU--GAGGUCCU-----GUGUAUCCCUG---CG .((((((((((((((..(....)..))))))))).)))-)).....(((.(((((...(.(((...--..))).).-----...))))).))---). ( -26.20) >DroSec_CAF1 31590 93 - 1 UACGUAUAUGGCAUAUUGCAUGUCCUAUGCCAUAUUGC-GCCAUUUGCAUGGAUGAGUGUCUCUAUCGGAGGUCCUUUUUGGUGUGCUCCUG---AG ..(((((((((((((..........))))))))).)))-)......((((.((.(((..((((.....))))..))).)).)))).......---.. ( -26.10) >DroSim_CAF1 31833 93 - 1 UACGUAUAUGGCAUAUUGCAUGUCCUAUGCCAUAUUGC-GCCAUUUGCAUGGAUGAGUGUCUCUAUCGGAGGUCCUUUUUGGUGUGCUCCUG---AG ..(((((((((((((..........))))))))).)))-)......((((.((.(((..((((.....))))..))).)).)))).......---.. ( -26.10) >DroEre_CAF1 32577 89 - 1 UACGUAUAUGGCAUAUUGCAUGUCCUAUGCCAUAUUGC-GCCAUUUGCAUGGAUGAGUGUCUCUAUCGGAGGUCCUUUUUGGUGC-------CUGUG ..(((((((((((((..........))))))))).)))-).(((..((((.((.(((..((((.....))))..))).)).))))-------..))) ( -29.20) >DroPer_CAF1 39781 87 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGCGGGCAUUUGCAUGGAUGAGUGUAUCUAU--GAGGUCCU-----GUGUAUCCCUG---CG ....................................(((((....(((((((((..(((....)))--...)))).-----)))))..))))---). ( -15.10) >consensus UACGUAUAUGGCAUAUUGCAUGUCCUAUGCCAUAUUGC_GCCAUUUGCAUGGAUGAGUGUCUCUAUCGGAGGUCCUUUUUGGUGUGCUCCUG___AG .....((((((((((..........))))))))))...........((((.((.(((..((((.....))))..))).)).))))............ (-15.68 = -17.07 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:40 2006