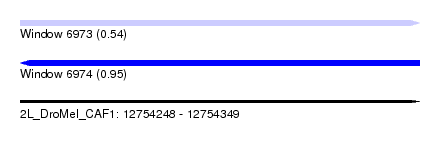
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,754,248 – 12,754,349 |
| Length | 101 |
| Max. P | 0.947903 |

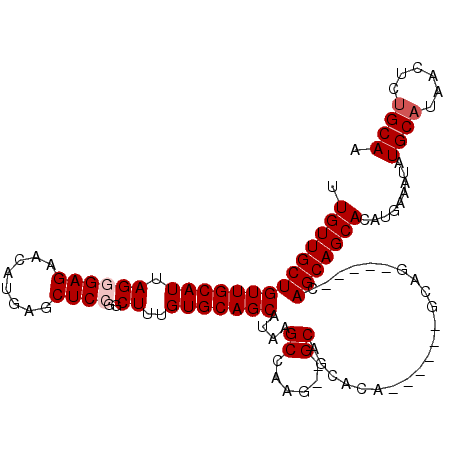
| Location | 12,754,248 – 12,754,349 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.536057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
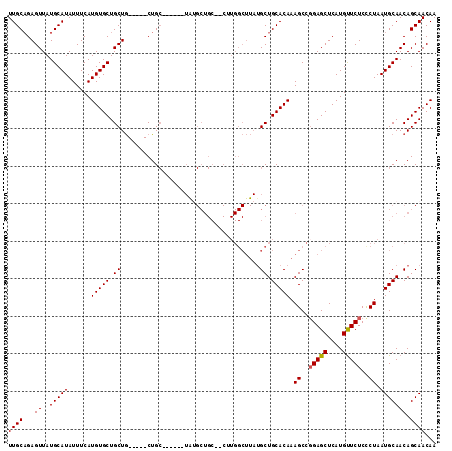
>2L_DroMel_CAF1 12754248 101 + 22407834 UUGCAGAGUUAUGCAUAUUUCAUGUGCUGCUG-----CUGC------UGUGCUGC--CUUGGCUUAUGCUGCACAAAGCCUGAGCUCAUGUUCUCCCUAAUGCAACAGCAACAA ..(((((((...((((((...)))))).))).-----))))------(((((((.--...((((((.(((......))).))))))..(((..........))).)))).))). ( -29.30) >DroSec_CAF1 31285 101 + 1 UUGCAGAGUUAUGCAUAUUUCAUGUGCUGCUG-----CUGC------UAUGCUGC--CUUGGCUUAUGCUGCACAAAGCCGGAGCUCAUGUUCUCCCUAAUGCAACAGCAACAA ..(((((((...((((((...)))))).))).-----))))------..(((((.--...(((....)))(((...((..(((((....)))))..))..)))..))))).... ( -29.40) >DroSim_CAF1 31538 106 + 1 UUGCAGAGUUAUGCAUAUUUCAUGUGCUGCUGCUAUGCUGC------UAUGCUGC--CUUGGCUUAUGCUGCACAAAGCCGGAGCUCAUGUUCUCCCUAAUGCAACAGCAACAA .((((......)))).....((((.(((.(.(((.((.(((------...((.((--....))....)).))))).))).).))).))))..........(((....))).... ( -28.70) >DroEre_CAF1 32319 83 + 1 UUGCAGAGUUAUGCAUAUUUCAUGUGCUGCUA-----CUG--------------------------AGCUGCACAAAGCCGGAGCUCAUGCUCUCCCUAAUGCAACAGCAACAA ((((...((..(((((......(((((.(((.-----...--------------------------))).))))).((..(((((....)))))..)).))))))).))))... ( -24.30) >DroYak_CAF1 31527 109 + 1 UUGCUGAGUUAUGCAUAUUUCAUGUGCUGCUG-----CUGCUGCUUCUGGGCUGCAGCUUGGCUUAUGCUGCACAAAGCCGGAGCUCAUGUUCUCUCUAAUGCAACAGCAACAA ((((((.....(((((......(((((....(-----((((.(((....))).)))))..(((....)))))))).((..(((((....)))))..)).))))).))))))... ( -36.80) >consensus UUGCAGAGUUAUGCAUAUUUCAUGUGCUGCUG_____CUGC______UAUGCUGC__CUUGGCUUAUGCUGCACAAAGCCGGAGCUCAUGUUCUCCCUAAUGCAACAGCAACAA ((((...((..(((((......(((((.((.....................................)).))))).((..(((((....)))))..)).))))))).))))... (-18.55 = -18.59 + 0.04)

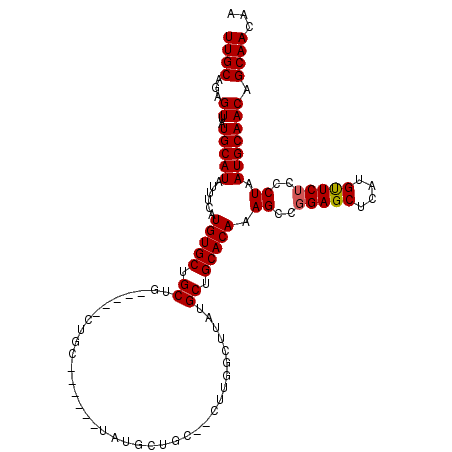
| Location | 12,754,248 – 12,754,349 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -22.52 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12754248 101 - 22407834 UUGUUGCUGUUGCAUUAGGGAGAACAUGAGCUCAGGCUUUGUGCAGCAUAAGCCAAG--GCAGCACA------GCAG-----CAGCAGCACAUGAAAUAUGCAUAACUCUGCAA .((((((((((((......(((........)))..((((((.((.......))))))--))......------))))-----)))))))).........((((......)))). ( -35.30) >DroSec_CAF1 31285 101 - 1 UUGUUGCUGUUGCAUUAGGGAGAACAUGAGCUCCGGCUUUGUGCAGCAUAAGCCAAG--GCAGCAUA------GCAG-----CAGCAGCACAUGAAAUAUGCAUAACUCUGCAA .((((((((((((.....((((........)))).((((((.((.......))))))--))......------))))-----)))))))).........((((......)))). ( -38.60) >DroSim_CAF1 31538 106 - 1 UUGUUGCUGUUGCAUUAGGGAGAACAUGAGCUCCGGCUUUGUGCAGCAUAAGCCAAG--GCAGCAUA------GCAGCAUAGCAGCAGCACAUGAAAUAUGCAUAACUCUGCAA .(((.((((((((.....((((........)))).(((((((((.((..........--)).)))))------).)))...))))))))))).......((((......)))). ( -35.10) >DroEre_CAF1 32319 83 - 1 UUGUUGCUGUUGCAUUAGGGAGAGCAUGAGCUCCGGCUUUGUGCAGCU--------------------------CAG-----UAGCAGCACAUGAAAUAUGCAUAACUCUGCAA .((((((((((((((.((((.((((....))))...))))))))))).--------------------------...-----.))))))).........((((......)))). ( -26.30) >DroYak_CAF1 31527 109 - 1 UUGUUGCUGUUGCAUUAGAGAGAACAUGAGCUCCGGCUUUGUGCAGCAUAAGCCAAGCUGCAGCCCAGAAGCAGCAG-----CAGCAGCACAUGAAAUAUGCAUAACUCAGCAA .((((((((((((....(.(((........)))).(((((.((((((.........)))))).....))))).))))-----)))))))).........(((........))). ( -37.90) >consensus UUGUUGCUGUUGCAUUAGGGAGAACAUGAGCUCCGGCUUUGUGCAGCAUAAGCCAAG__GCAGCACA______GCAG_____CAGCAGCACAUGAAAUAUGCAUAACUCUGCAA .((((((((((((((.((((((........))))..))..)))))))....((......))......................))))))).........((((......)))). (-22.52 = -23.22 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:38 2006