| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,749,482 – 12,749,598 |
| Length | 116 |
| Max. P | 0.684953 |

| Location | 12,749,482 – 12,749,598 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -18.03 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
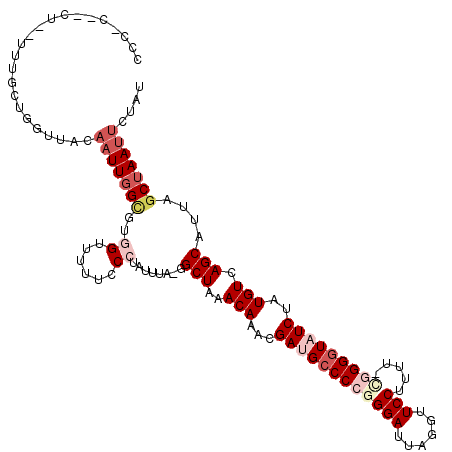
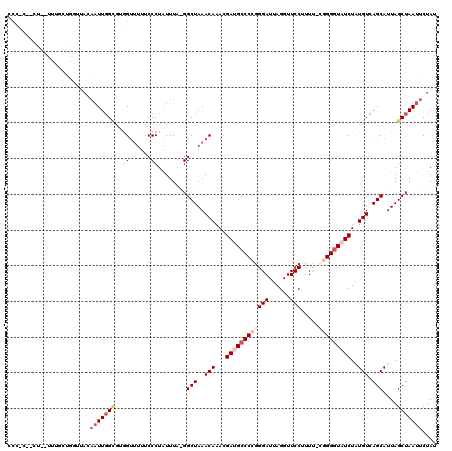
>2L_DroMel_CAF1 12749482 116 + 22407834 CCCCC--CUUUUUUUCUGAUUACAAUUGGCGUGGUUUUUCCCUAUUUA-GGCUAAACAAACGAAGCCCCGGGAUUAGGUUCCUUUU-CGGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU .....--....................((((((((((...(((....)-))..))))....((.((((((..(..((....)))..-)))))).))))))))(((....)))........ ( -27.00) >DroPse_CAF1 29223 119 + 1 CCAACUUCUCUUUUCUUGGCUACAAUUGGUCUUGGUGGUCCCUGUUUGGGGCUAAACAAACGAUGCCCUGGGAUUAGGUUCCAUUC-UUGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU ...............(((((((...(((((.(((.(((((((.....)))))))..)))))(((((((.(((((........))))-).))))))).....)))...)))))))...... ( -33.20) >DroSec_CAF1 26436 102 + 1 CCC-----------------CACAAUUGGCGUGGUUUUUCCCUAUUUA-GGCUAAACAAACGAUGCCCCGGGAUUAGGUUCCUUUUCGGGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU ...-----------------.......((((((((((...(((....)-))..))))....((((((((.(((..((....)).))).))))))))))))))(((....)))........ ( -30.20) >DroSim_CAF1 26777 116 + 1 CCCAC--UUACUCUGCUGGUUACAAUUGGCGUGGUUUUUCCCUAUUUA-GGCUAAACAAACGAUGCCCCGGGAUUAGGUUCCUUUU-GGGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU .....--.........((((((...((((((((((((...(((....)-))..))))....((((((((((((......))))...-.))))))))))))))))...))))))....... ( -32.30) >DroYak_CAF1 26989 116 + 1 CCCGC--CCACUUUGCUGGUUACAAUUGGCGCGGUUUUUCCCUAUUUA-GGCUAAACAAACGAUGCCCCGGGAUUAGGUUCCGUUU-CGGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU .((((--(((..(((.......))).))).))))..............-((((((......(((((((((..((........))..-)))))))))..((....))))))))........ ( -35.90) >DroAna_CAF1 25742 96 + 1 --------------UCUUAU-----UUCG---AGUCUUGCCCUAUUUA-GGCUAAACAAACGACGGCCCGGGAUUAGGUUCCAUUC-CCGGGUAUCUGUGUCAGCAUUAGCUAAUUCUAU --------------......-----....---.((...(((.......-)))...))....((((((((((((...(....)..))-)))))).....))))(((....)))........ ( -24.30) >consensus CCC_C__CU__UUUGCUGGUUACAAUUGGCGUGGUUUUUCCCUAUUUA_GGCUAAACAAACGAUGCCCCGGGAUUAGGUUCCUUUU_CGGGGUAUCUAUGUCAGCAUUAGCUAAUUCUAU .......................(((((((..((......))........(((..(((...((((((((((((......))).....)))))))))..))).)))....))))))).... (-18.03 = -20.00 + 1.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:35 2006