
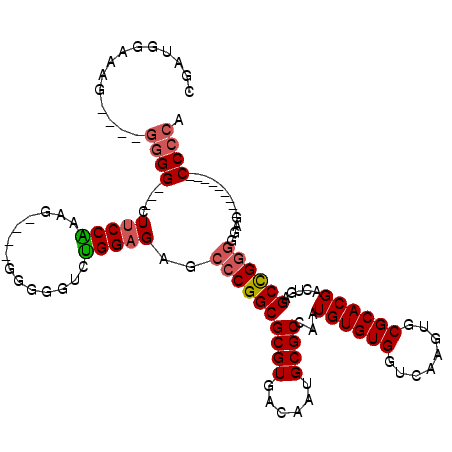
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,748,302 – 12,748,515 |
| Length | 213 |
| Max. P | 0.772846 |

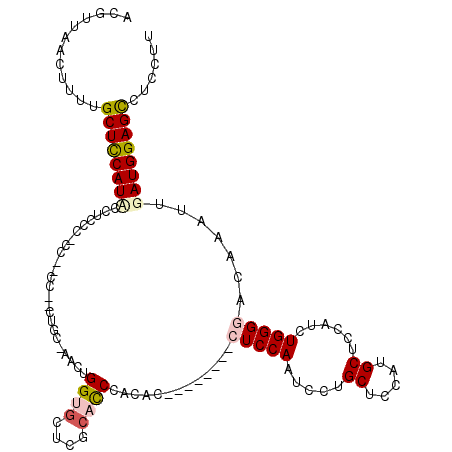
| Location | 12,748,302 – 12,748,404 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.99 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12748302 102 + 22407834 CGAGGGAAAG----GGGGUUGCUUCCGAAG-----GGGGUCUGGAGAGCCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGGGGGAG---------CCCCA ..........----((((((.((((((...-----(((.((....)).)))(((((((....)))))))..((((((........)))))).......))))))))---------)))). ( -46.40) >DroSec_CAF1 25263 106 + 1 CGAUGGAAAG----CGGGUU-CUUCCAAAGGAGGGGGGGUCUGGAGUGCCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGGGGGAG---------CCCCA .........(----.(((((-(((((...((.((..((((.......))))(((((((....)))))))..((((((........)))))).))...)))))))))---------)))). ( -42.70) >DroSim_CAF1 25641 102 + 1 CGAUGGAAAG----GGGGUU-CUUCCAAAG----GGAGGUCUGGAGUGCCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGGGGGAG---------CCCCA ..........----((((((-(((((....----((.((((.((.....))(((((((....)))))))...(((((........)))))))))...)))))))))---------)))). ( -45.90) >DroEre_CAF1 26795 108 + 1 CGACGGCGAG----GGGG---AUUCCGAAG----G-GGGUCUGGAGAGGCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCUGGGGGAGCUGGGGCAGCCCCA ...((((..(----(.((---(((((....----)-))))))((.....))(((((((....)))))))..((((((........)))))).))..)))).(((.((((...))))))). ( -47.00) >DroYak_CAF1 25786 113 + 1 AGGUGGGAAGGGGGGGGG---GUUCCAAAG----GGGGGUAUGGAGAGCCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGGGGGAGCCGGGGGAUCCCCA ..............((((---(((((....----((((((.......))))(((((((....)))))))..((((((........)))))).))...((((.....))))))))))))). ( -48.00) >DroAna_CAF1 24711 100 + 1 CUUGAGGAUG----GCGG---GUGCCGGCU----G-G----CGGAGAACCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGAGGAAGCCGGG----CCCCA ..........----..((---(..((((((----(-(----(..((.....(((((((....)))))))..((((((........)))))).))..))).....)))))).----.))). ( -42.60) >consensus CGAUGGAAAG____GGGG___CUUCCAAAG____GGGGGUCUGGAGAGCCCGGCGCGUGACAAUGCGCCAAUGUGUGGUCAAGUGCGCACGACUGAGCCGGGGGAG_________CCCCA ..............((((....(((((..............)))))..((((((((((......))))...((((((........)))))).....)))))).............)))). (-29.68 = -29.99 + 0.31)

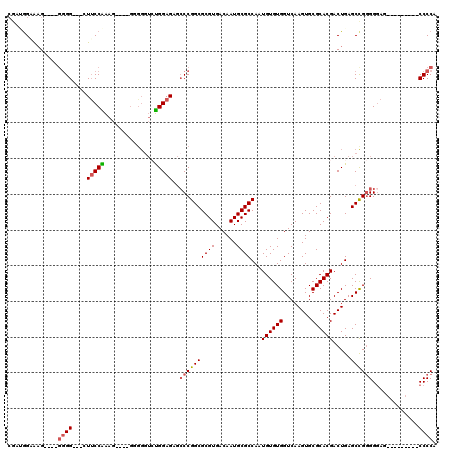
| Location | 12,748,373 – 12,748,476 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -22.64 |
| Energy contribution | -24.08 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12748373 103 - 22407834 AACUGGUGCUCGCACCCACAC--------CUCCAAUCCUGCUCCAUGCUCCAUCUGGGAACAAAUUGAUGGAGUCUCCUUUGGGG---------CUCCCCCGGCUCAGUCGUGCGCACUU ....(((((..((((......--------..........((((((.((((((((((....))....))))))))......)))))---------)......(((...)))))))))))). ( -31.70) >DroSec_CAF1 25338 103 - 1 AACUGGUGCUCGAACCCACAC--------CUCCAAUCCUGCUCCACGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUUUGGGG---------CUCCCCCGGCUCAGUCGUGCGCACUU .....((((.(((........--------........(((..((..((((((((((....))....)))))))).......((((---------...))))))..)))))).)))).... ( -33.59) >DroSim_CAF1 25712 103 - 1 AACUGGUGCUCGCACCCACAC--------CUCCAAUCCUGCUCCACGCUCCAUCUGGGAACAAAUUGAUGGAGCCUCCUUUGGGG---------CUCCCCCGGCUCAGUCGUGCGCACUU ....((((....)))).....--------.........(((..((((((....(((((..(.....)..((((((((....))))---------)))))))))...)).)))).)))... ( -32.70) >DroEre_CAF1 26863 110 - 1 UACGGGUGCCCGCCUCCACAC--------CUCCAAUCCUGC--CGUGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUUUGGGGCUGCCCCAGCUCCCCCAGCUCAGUCGUGCGCACUU ...((((((.(((..(.....--------............--.(.((((((((((....))....)))))))))....((((((..((....))..))))))....)..))).)))))) ( -36.90) >DroYak_CAF1 25859 118 - 1 UACGGGUGCCCGCCCCCACACCUCCAUACCUCCAAUCCUGC--CGUGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUUUGGGGAUCCCCCGGCUCCCCCGGCUCAGUCGUGCGCACUU (((((((....))))........................((--((.((((((((((....))....)))))))).......(((((.((...)).)))))))))......)))....... ( -38.00) >DroAna_CAF1 24775 92 - 1 -----ACCCCCGGCCCCAUAGA-------------------CACACACACACUCGGGGGACAAAUUGAUGGAGCCUCCUUUGGGG----CCCGGCUUCCUCGGCUCAGUCGUGCGCACUU -----...((.(((((((..((-------------------...........))(((((.((......))...)))))..)))))----)).))........((.((....)).)).... ( -25.90) >consensus AACUGGUGCCCGCACCCACAC________CUCCAAUCCUGCUCCACGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUUUGGGG_________CUCCCCCGGCUCAGUCGUGCGCACUU ....(((((.(((.................................((((((((((....))....)))))))).....((((((............)))))).......))).))))). (-22.64 = -24.08 + 1.45)


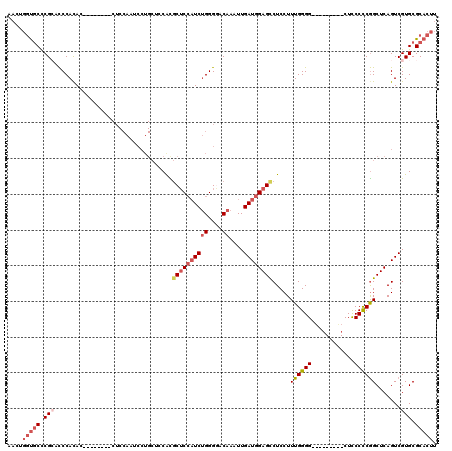
| Location | 12,748,404 – 12,748,515 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12748404 111 - 22407834 ACGUUAACUUUUGCUCCAUACCUCCCCCCGUCCCCCAGC-AACUGGUGCUCGCACCCACAC--------CUCCAAUCCUGCUCCAUGCUCCAUCUGGGAACAAAUUGAUGGAGUCUCCUU ............(((((((..............(((((.-....((((....)))).....--------..........((.....)).....)))))..(.....))))))))...... ( -21.40) >DroSec_CAF1 25369 107 - 1 ACGUUAACUUUUGCUCCAUACCGCCCUCC--CC--UUGC-AACUGGUGCUCGAACCCACAC--------CUCCAAUCCUGCUCCACGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUU ............(((((((.......(((--((--.((.-...(((.((..((........--------......))..)).))).....))...))))).......)))))))...... ( -25.18) >DroSim_CAF1 25743 109 - 1 ACGUUAACUUUUGCUUCAUCCAACCCACC--CCCUCUGC-AACUGGUGCUCGCACCCACAC--------CUCCAAUCCUGCUCCACGCUCCAUCUGGGAACAAAUUGAUGGAGCCUCCUU ..........((((...............--......))-))..((((....)))).....--------.................((((((((((....))....))))))))...... ( -20.70) >DroEre_CAF1 26903 98 - 1 ACGUUAACUUUUGCUCCAUCCCC------------CUGCUUACGGGUGCCCGCCUCCACAC--------CUCCAAUCCUGC--CGUGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUU ............((((((((...------------........(((((..........)))--------))..........--.((.((((....)))))).....))))))))...... ( -23.90) >DroYak_CAF1 25899 102 - 1 ACGUUAACUUUUGCUCCAUUCCU------------C----UACGGGUGCCCGCCCCCACACCUCCAUACCUCCAAUCCUGC--CGUGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUU ............((((((((...------------.----...((((....))))..........................--.((.((((....)))))).....))))))))...... ( -23.40) >consensus ACGUUAACUUUUGCUCCAUACCUCCC_CC__CC__CUGC_AACUGGUGCUCGCACCCACAC________CUCCAAUCCUGCUCCAUGCUCCAUCUGGGGACAAAUUGAUGGAGCCUCCUU ............((((((((........................((((....)))).............(((((.....((.....))......))))).......))))))))...... (-16.66 = -17.50 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:34 2006