
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,744,882 – 12,744,979 |
| Length | 97 |
| Max. P | 0.978184 |

| Location | 12,744,882 – 12,744,979 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12744882 97 - 22407834 AUAUUAGUCAGGCAAAAUG-UGAGGCUACCAGCUUUAAGUUGUCAAAAAAGAAA----AGA-AAUUCGAAGGAUUUCGAAGAUGACAAUACAUUUAAAGCUAA- ....(((((..((.....)-)..)))))..(((((((((((((((.........----.((-((((.....)))))).....)))))))....))))))))..- ( -21.16) >DroSec_CAF1 21827 102 - 1 AUAUUAGUCAGGCAAAAUG-UGAGGCUACUAGCUUUAAGUUGUCAAAAAAGAAAGUAAAGAAAGUUCGAAGGAUUUCAAAGAUGACAACACAUUUAAAGCUAA- ....(((((..((.....)-)..))))).((((((((((((((((.....((((.....((....))......)))).....)))))))....))))))))).- ( -22.50) >DroSim_CAF1 22187 102 - 1 AUAUUAGUCAGGCAAAAUG-UGAGGCUACGAGCUUUAAGUUGUCAAAAAAGAAAGUAAAGAAAGUUCGAAGGAUUUCGAAGAUGACAACACAUUUAAAGCUAA- ....(((((..((.....)-)..)))))..(((((((((((((((.....((((.....((....))......)))).....)))))))....))))))))..- ( -22.50) >DroEre_CAF1 23557 100 - 1 AUAUUAGUCAGGCAAAAUG-UGAGGCUAA-GGCUUUAAGUUGUCAAAAG--CCAAGCAAAAAGGUUCGAAGGAUUUCCAAGAUGACAACACAUUUAAAGCUAAA ...((((((..((.....)-)..))))))-(((((((((((((((...(--(...)).....((.((....))...))....)))))))....))))))))... ( -25.80) >DroYak_CAF1 22399 100 - 1 AUUUUAGUCAGGCAAAAUG-UGAGGCUAA-AGCUUUAAGUUGUCAAAAG--CCACGCAAGAAGGUUCGAAGGAUUUCUAAGAUGACAACACAUUUAAAGCUAAA ...((((((..((.....)-)..))))))-(((((((((((((((...(--((.(....)..)))((....)).........)))))))....))))))))... ( -26.20) >DroAna_CAF1 21509 88 - 1 A---CAGUCAGCCAGAGUUUUGAGAAUU-AAGCUUAAAGCUGUCAAAUG--CAAGCAUCUGAAGUUCGAAGGAUUUCUGAGAUGACAACACU---------AU- .---..((((.((((((((((..(((((-.(((.....))).(((.(((--....))).))))))))..)))).))))).).))))......---------..- ( -20.80) >consensus AUAUUAGUCAGGCAAAAUG_UGAGGCUAC_AGCUUUAAGUUGUCAAAAA__AAAGGAAAGAAAGUUCGAAGGAUUUCGAAGAUGACAACACAUUUAAAGCUAA_ ......(((((((((.....((((((.....))))))..))))).....................((....)).........)))).................. (-13.48 = -13.87 + 0.39)

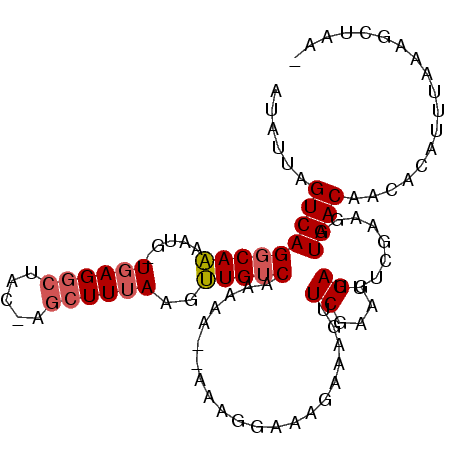

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:30 2006