
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,705,570 – 12,705,684 |
| Length | 114 |
| Max. P | 0.500000 |

| Location | 12,705,570 – 12,705,684 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12705570 114 + 22407834 GCUCU-----GUUUAUUUC-UUACAGUGGGUGUGGGAGAAGCGGCCAACUUUGCGGCCUACGCCUUUGCCCCCGCUUCAUUGGUCACUCCGUUGGGCGCGCUGAGUGUUAUCAUCUCAGC .....-----.....((((-(((((.....))))))))).(((.(((((...((((.....((....))..))))......((.....))))))).)))((((((((....)).)))))) ( -37.50) >DroVir_CAF1 1208 119 + 1 GUUAUCGUUUGUGUGUGCC-UUGCAGUGGGCUUGGGCGAAGCAGCCAAUUUUGCCGCGUAUGCAUUCGCACCCGCCUCCCUGGUGACGCCGCUGGGCGCGCUGAGCGUCAUCAUCUCGGC (....((((((.(((((((-...((((((((...(((......)))......)))((((.(((....)))..((((.....))))))))))))))))))))))))))....)........ ( -43.60) >DroPse_CAF1 338 113 + 1 GUCAU-----CAUU-GUCU-UUGCAGUGGGCCUGGGUGAGGCGGCUAACUUUGCCGCCUACGCCUUUGCGCCCGCUUCCCUGGUGACCCCCCUGGGGGCACUGAGUGUGAUUAUUUCGUC ((((.-----((((-....-....((((((((.((((((((((((.......))))))).)))))..).))))))).....((((.((((....)))))))).))))))))......... ( -52.90) >DroGri_CAF1 430 112 + 1 GAUAUC-------UGUUUU-UUGCAGUGGGUCUUGGCGAGGCAGCCAAUUUUACGGCGUAUGCGUUUGCUCCCGCCUCGCUGGUGACGCCCCUGGGCGCACUGAGUGUCAUCAUUUCGGC (((((.-------......-...(((((.((((..(((((((.(((........)))(((......)))....)))))))..).)))(((....))).))))).)))))........... ( -41.80) >DroWil_CAF1 745 115 + 1 AAUCA-----AUUUCUUUCUUUACAGUGGGACUGGGUGAGGCGGCCAACUUCGCUGCCUAUGCCUUUGCUCCGGCGUCUCUCGUCACACCUCUGGGUGCUCUCAGUGUUAUCAUAUCAGC .....-----............(((.(((((..((((((((((((.......))))))).))))).......((((.....)))).((((....)))).))))).)))............ ( -32.30) >DroPer_CAF1 338 113 + 1 AUCAU-----CAUU-GUCU-UUGCAGUGGGCCUGGGUGAGGCGGCUAACUUUGCCGCCUACGCCUUUGCGCCCGCUUCCCUGGUGACCCCCCUGGGGGCACUGAGUGUGAUUAUUUCGUC ...((-----(((.-.((.-....((((((((.((((((((((((.......))))))).)))))..).))))))).....((((.((((....))))))))))..)))))......... ( -52.60) >consensus GUUAU_____CUUU_UUCC_UUGCAGUGGGCCUGGGUGAGGCGGCCAACUUUGCCGCCUACGCCUUUGCGCCCGCUUCCCUGGUGACCCCCCUGGGCGCACUGAGUGUCAUCAUUUCGGC .....................(((.(((((...(((((..((.((.......)).))...))))).....)))))......((((.((((....))))))))..)))............. (-22.20 = -22.65 + 0.45)
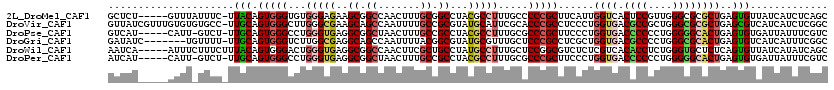
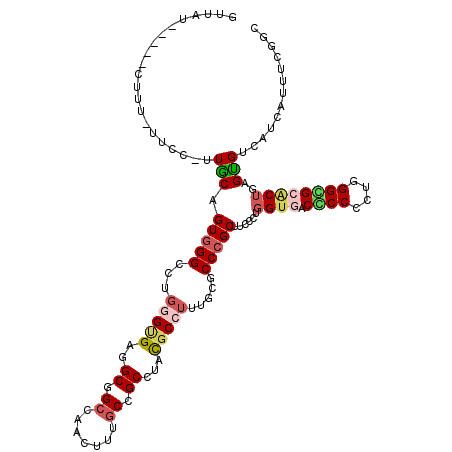
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:22 2006