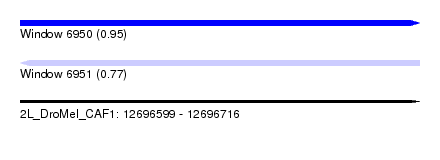
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,696,599 – 12,696,716 |
| Length | 117 |
| Max. P | 0.950595 |

| Location | 12,696,599 – 12,696,716 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -30.64 |
| Energy contribution | -32.03 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
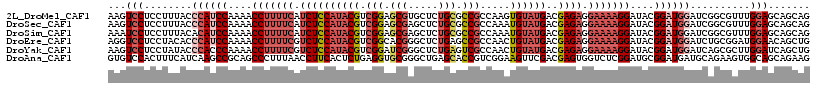
>2L_DroMel_CAF1 12696599 117 + 22407834 CUGCUGCUCCAAACGCCGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACACUUGGCGGCGCAGAGCACGCUCCGACGUAUGGAGAUGAAAAGGUUUUGGAUGGGUAAAGGAGGACUU .......(((.....((.(((((((((...(((((((.((((..(((((..((((.((((.......)))))))).))))))))).)))))))...)))))))))...)).)))... ( -44.20) >DroSec_CAF1 335 117 + 1 CUGCUGCUCCAAACGCCGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACAUUUGGCGGCGCAGAGCUCGCUCCGACGUAUGGAGAUGAAAAGGUUUUGGAUGGGUAAAGGAGGACUU .......(((.....((.(((((((((...(((((((.((((..(((((.((.((((((.....)).))).).)).))))))))).)))))))...)))))))))...)).)))... ( -44.30) >DroSim_CAF1 335 117 + 1 CUGCUGCUCCAAACGCCGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACAUUUGGCGGCGCAGAGCUCGCUCCGACGUAUGGAGAUGAAAAGGUUUUGGAUGUGUAAAGGAGGAUUU .......(((.....((.(..((((((...(((((((.((((..(((((.((.((((((.....)).))).).)).))))))))).)))))))...))))))..)...)).)))... ( -38.70) >DroEre_CAF1 304 117 + 1 CAGCUGUUCCAUCCGCAGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACAGUUGGCGGCUCAGAGCCCGUGCCGACGUAUGGAGACGAAAAGGUUUUGGAUGGGUGUAGGAGGACCU .......(((.(((..(.(((((((((...(((((((.((((..(((((.(((((((((.....)))...))))))))))))))).)))))))...))))))))).).))))))... ( -54.10) >DroYak_CAF1 342 117 + 1 CAGCUGAUCCAAGCGCUGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACAGUUGGCGACUCAGAGCCCGAUCCGACGUAUGGAGACGAAAAGGUUUUGGGUGGGUAUAGGAGGACUU ..(((......))).((((((((((((...(((((((.((((..(((((.(((((((..........))..)))))))))))))).)))))))...))))))))).)))........ ( -41.70) >DroAna_CAF1 701 117 + 1 CUUCUGCUGCCACUUCUGCAUCAUCCGCAUCCGAGACCACUCGUCGAACUUCCGACGGUGCUCAGCCCGCACCUCAGAGUGAAGGUUAAAGGGCUGCGGCUUGAUGAAAGUGGACAC .......((((((((...(((((.((((((((..((((((((((((......))))(((((.......)))))...))))...))))...))).)))))..))))).)))))).)). ( -47.60) >consensus CUGCUGCUCCAAACGCCGAUCCAUCCGUAUCCUUUUCCUCUCGUCAUACAUUUGGCGGCGCAGAGCCCGCUCCGACGUAUGGAGAUGAAAAGGUUUUGGAUGGGUAAAGGAGGACUU .......(((.....((.(((((((((...(((((((.((((((((......))))((.((.......)).))........)))).)))))))...)))))))))...)).)))... (-30.64 = -32.03 + 1.39)



| Location | 12,696,599 – 12,696,716 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -24.93 |
| Energy contribution | -27.15 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12696599 117 - 22407834 AAGUCCUCCUUUACCCAUCCAAAACCUUUUCAUCUCCAUACGUCGGAGCGUGCUCUGCGCCGCCAAGUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCGGCGUUUGGAGCAGCAG ..((.((((...((((((((....(((((((.((((((((((...(.(((.((.....))))))...))))))..)))).)))))))....))))))......))..))))..)).. ( -39.30) >DroSec_CAF1 335 117 - 1 AAGUCCUCCUUUACCCAUCCAAAACCUUUUCAUCUCCAUACGUCGGAGCGAGCUCUGCGCCGCCAAAUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCGGCGUUUGGAGCAGCAG ..((.((((...((((((((....(((((((.(((((((((((..(.(((.((.....))))))..)))))))..)))).)))))))....))))))......))..))))..)).. ( -41.70) >DroSim_CAF1 335 117 - 1 AAAUCCUCCUUUACACAUCCAAAACCUUUUCAUCUCCAUACGUCGGAGCGAGCUCUGCGCCGCCAAAUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCGGCGUUUGGAGCAGCAG .....((((...((.(((((....(((((((.(((((((((((..(.(((.((.....))))))..)))))))..)))).)))))))....)))))(.....)))..))))...... ( -38.70) >DroEre_CAF1 304 117 - 1 AGGUCCUCCUACACCCAUCCAAAACCUUUUCGUCUCCAUACGUCGGCACGGGCUCUGAGCCGCCAACUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCUGCGGAUGGAACAGCUG .(.((((((.....((((((....(((((((.(((((((((((.(((...(((.....)))))).)).)))))..)))).)))))))....))))))......))).))).)..... ( -45.60) >DroYak_CAF1 342 117 - 1 AAGUCCUCCUAUACCCACCCAAAACCUUUUCGUCUCCAUACGUCGGAUCGGGCUCUGAGUCGCCAACUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCAGCGCUUGGAUCAGCUG ..............(((.((....(((((((.(((((((((((.((.((((...))))....)).)).)))))..)))).)))))))....)).)))..((((..........)))) ( -33.80) >DroAna_CAF1 701 117 - 1 GUGUCCACUUUCAUCAAGCCGCAGCCCUUUAACCUUCACUCUGAGGUGCGGGCUGAGCACCGUCGGAAGUUCGACGAGUGGUCUCGGAUGCGGAUGAUGCAGAAGUGGCAGCAGAAG ((..((((((((((((..(((((((((....(((((......)))))..)))))((.(((((((((....)))))).))).))......)))).)))))..)))))))..))..... ( -49.90) >consensus AAGUCCUCCUUUACCCAUCCAAAACCUUUUCAUCUCCAUACGUCGGAGCGGGCUCUGCGCCGCCAAAUGUAUGACGAGAGGAAAAGGAUACGGAUGGAUCGGCGUUUGGAGCAGCAG ...(((........((((((....(((((((.((((((((((.(((.(((.....))).))).....))))))..)))).)))))))....))))))..........)))....... (-24.93 = -27.15 + 2.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:15 2006