
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,686,757 – 12,686,856 |
| Length | 99 |
| Max. P | 0.748992 |

| Location | 12,686,757 – 12,686,856 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.94 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12686757 99 - 22407834 AAGUGAUUUCUUAUCCAACAAAUCCACACUCGACAACCAA-AGCAACCAAAAAAAUAA----A-AAAAAA-AGGGGUUGCUGGUUUGGGUCCUUCCUUCGCAUCCU--- ..((((......((((((.................((((.-.((((((..........----.-......-...)))))))))))))))).......)))).....--- ( -17.29) >DroSec_CAF1 67276 97 - 1 AAGUGAUUUCUUAUCCAACA-ACCCACACUCGCCAACCAA-CGCAACAAACAAAGAAA---AAAAAGAAACGGGGGUUACUGGUGUGGGUCC----UUCGCAUCCU--- ..((((..............-(((((((((.(..((((..-((...............---.........))..)))).).)))))))))..----.)))).....--- ( -21.39) >DroSim_CAF1 69233 100 - 1 AAGUGAUUUCUUAUCCAACAAAUCCACACUCGACAACCAA-AGCAACAAAAAAAAAAAAUAAAAAAGAAA-AGGGGUUGCUGGUGUGGGUCC----UUCGCAUCCU--- ..((((...............(((((((((.(.(((((..-.............................-...)))))).)))))))))..----.)))).....--- ( -18.85) >DroEre_CAF1 69176 89 - 1 AAGUGAUUUCUUAUCCAGCAAAUCCACACUCGACAACCAAGCCGAACAUG----G-----------AAAA-AGGGGUUGCUGGUGUGGGUCC----UUCGCAUCCUUCG ..((((...((((((((((((..((.(....)....(((.(.....).))----)-----------....-.))..))))))).)))))...----.))))........ ( -22.40) >DroYak_CAF1 70093 84 - 1 AAGUGAUUUCUUAUCCAGCAAAUCCACACUCGACAACCAA-CUGAACAUA----------------AAAA-AGGGGUUGCUGGUGUGGGUCC----UUCGCAUCCU--- ..((((...............(((((((((.(.(((((..-((.......----------------....-)).)))))).)))))))))..----.)))).....--- ( -20.47) >DroAna_CAF1 58782 86 - 1 AAGUGAUUUCUUAUCCAACAAAUCCACACUCGACAAGCAA------CAAACAACG---------AGGAAA-AGGGGUUGCUGGUGUGGGUUC----UUGGCAUCCU--- ..............((((..((((((((((.....(((((------(...(....---------......-.)..)))))))))))))))).----))))......--- ( -21.40) >consensus AAGUGAUUUCUUAUCCAACAAAUCCACACUCGACAACCAA_CGCAACAAA_AAAG_________AAAAAA_AGGGGUUGCUGGUGUGGGUCC____UUCGCAUCCU___ ..((((...............(((((((((.(.(((((....................................)))))).))))))))).......))))........ (-15.41 = -15.94 + 0.53)

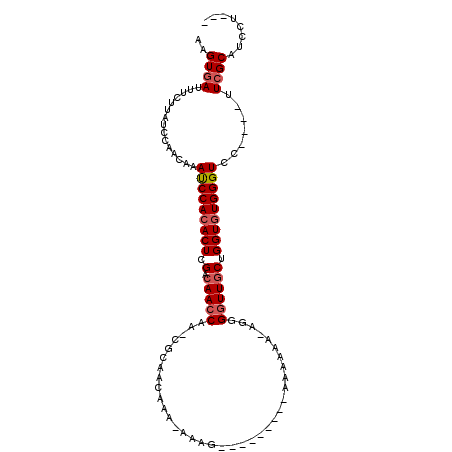

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:09 2006