| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,675,333 – 12,675,444 |
| Length | 111 |
| Max. P | 0.979765 |

| Location | 12,675,333 – 12,675,444 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.01 |
| Energy contribution | -17.15 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
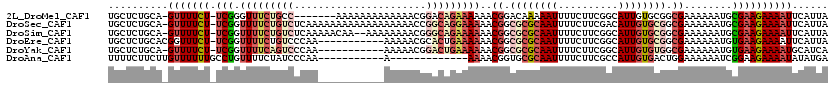
>2L_DroMel_CAF1 12675333 111 - 22407834 UGCUCUGCA-GUUUUCU-UCGGGUUUCUGCC-------AAAAAAAAAAAAACGGACAGAAAAAACGGACAAAAAUUUUCUUCGGCAUUGUGCGGCGAAAAAAUGCGAAGAAAAUUCAUUA .....((.(-(((((((-(((.((((.((((-------.............(((..((((((............)))))))))((.....))))))...)))).)))))))))))))... ( -25.40) >DroSec_CAF1 56007 118 - 1 UGCUCUGCA-GUUUUCU-UCGGUUUUCUGUCUCAAAAAAAAAAAAAAAAAACCGGCAGGAAAAACGGCGCGCAAUUUUCUUCGACAUUGUGCGGCGAAAAAAUGCGAAGAAAAUUCAUUA .....((.(-(((((((-(((.(((((((((......................)))))))))..((.((((((((.((....)).)))))))).))........)))))))))))))... ( -34.05) >DroSim_CAF1 57901 116 - 1 UGCUCUGCA-GUUUUCU-UCGGUUUUCUGUCUCAAAAACAA--AAAAAAAACGGGCAGAAAAAACGGCGCGCAAUUUUCUUCGGCAUUGUGCGGCGAAAAAAUGCGAAGAAAAUUCAUUA .....((.(-(((((((-(((.((((((((((.........--.........))))))))))..((.((((((((...(....).)))))))).))........)))))))))))))... ( -37.27) >DroEre_CAF1 57801 108 - 1 UGCUCUGCACGGUUUCU-UCGGUUUUCUGUCCCAA-----------AAAAACGCACUGAAAAAACGGCGCGCAAUUUUCUUCGGCAUUGUGCGGCGAAAAAAUGUGAAGAAAAUUCAUUA ...(((.((((((((.(-(((((............-----------........)))))).))))(.((((((((...(....).)))))))).).......)))).))).......... ( -25.05) >DroYak_CAF1 58579 107 - 1 UGCUCUGCA-GUUUUCU-UCGGUUUUCAGUCCCAA-----------AAAAACGGACUGAAAAAACGGCGCGCAAUUUUCUUCGGCAUUGUGUGGCGAAAAAAUGUGAAGAAAAUGCAUCA .....((((-.((((((-(((.((((((((((...-----------......))))))))))..((.((((((((...(....).)))))))).))........)))))))))))))... ( -38.90) >DroAna_CAF1 48385 96 - 1 UUUUCUUCUUGUUUUUUGCCUGUUUUCUAUCCCAA-----------A-------------AAAACGGUGCGCAAUUUUCUUCGCCAUUGUGACUGGAAAAAAUCGGAAGAAAAUAUAUGA (((((((((.((((((((((((((((.........-----------.-------------))))))).))..........((((....))))....))))))).)))))))))....... ( -21.30) >consensus UGCUCUGCA_GUUUUCU_UCGGUUUUCUGUCCCAA___________AAAAACGGACAGAAAAAACGGCGCGCAAUUUUCUUCGGCAUUGUGCGGCGAAAAAAUGCGAAGAAAAUUCAUUA ..........(((((((.(((.(((((((((......................)))))))))..((.((((((((..........)))))))).))........))))))))))...... (-15.01 = -17.15 + 2.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:07 2006