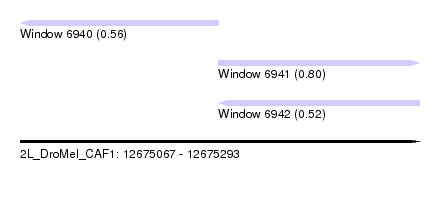
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,675,067 – 12,675,293 |
| Length | 226 |
| Max. P | 0.796874 |

| Location | 12,675,067 – 12,675,179 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12675067 112 - 22407834 GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCUCCUCUUCCUCUAUUGAUUUCCGUUGCUGAGGUUGCCUCAUCGAUUUGCGCUCGCUUUAAUCGGGUUCCUUUCUACU-----U--- ((((((...((((.....((((((.((.....))))))))))))......))))))((....((((..(((....(((((.((....))...)))))))).))))...)).-----.--- ( -31.80) >DroSec_CAF1 55740 113 - 1 GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCUCCUCUUCCUCUAUUGAUUUCCGUUGCUGAGGUUUCCUCAUCGAUUUGCGCUCGGUUUAAUCGGGUUCCUUUCUACU----AU--- ...((((((((((.....((((((.((.....))))))))..(((.(((((...(((.((((((((...))))).......)))..))).))))).))).)))))).))))----..--- ( -31.81) >DroSim_CAF1 57628 113 - 1 GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCUCCUCUUCCUCUAUUGAUUUCCGUUGCUGAGGUUUCCUCAUCGAUUUGCGCUCUCUUUAAUCGGGUUCCUUUCUACU----AU--- ...((((((((((.....((((((.((.....))))))))..(((.(((((....(((((.(((((...))))).))....)))......))))).))).)))))).))))----..--- ( -27.60) >DroEre_CAF1 57538 110 - 1 GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCU---CUUCCUCUAUUGAUUUCCGUUGCUGAGGUUUCCUCAUCGAUUUGCGCUCGCUUUAAUCGGGUUCCUUUCUACU----AU--- ...((((((((((((..(((((((.((.....)).---..))))))).))....((.....(((((...))))).(((((.((....))...))))))).)))))).))))----..--- ( -32.30) >DroYak_CAF1 58316 110 - 1 GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCU---CUUCCUCUAUUGAUUUCCGUUGCUGAGGUUUCCUCAUCGAUUUGCGCUCGCUUUAAUCGCGUUUCUUUCUACU----AC--- (((((.(((.(((.((((((((((.((.....)).---..)))))))..))).)))...(((((((...)))))..((((.((....))...)))))).))))))))....----..--- ( -29.80) >DroAna_CAF1 48116 119 - 1 GGAAGUAAAAGGACAUUUGGAGGCUGCUUU-UGUCCUUUUUGCUCUAUUGAUUUCCGUUACUGAGGUUUCCUCAUCGAUUUGCACUCGCUUUAAUCGGGAACCUUCCUCUUUUUUAUUUC (((((.(((((((((...((......))..-)))))))))............((((.....(((((...))))).(((((.((....))...))))))))).)))))............. ( -30.30) >consensus GGAAGUAAAAGGACAUUUAGAGGAUGCUUUGUGCUCCUCUUCCUCUAUUGAUUUCCGUUGCUGAGGUUUCCUCAUCGAUUUGCGCUCGCUUUAAUCGGGUUCCUUUCUACU____AU___ (((((.....(((.((((((((((.((.....))......)))))))..))).))).....(((((...)))))((((((.((....))...))))))....)))))............. (-24.38 = -24.80 + 0.42)


| Location | 12,675,179 – 12,675,293 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -26.89 |
| Energy contribution | -27.95 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12675179 114 + 22407834 GCAUUUGUAGCGCCGCAUCUU------CUGCCCCUCGUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCGCUGUCAAUCGGCGCUGCAAC ....(((((((((((.....(------(..(.....)..))................((((((..(((.(((((....)))))((((...))))...)))..))))))))))))))))). ( -35.80) >DroSec_CAF1 55853 114 + 1 GCAUUUGUAGCGCCGCAUCUU------CUGCCCCUCGUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCGCUGACAAUCGGCGCUGCAAC ....(((((((((((.((..(------(.((....(((.(((.....))).)))......(((((....(((((....)))))((((...))))))))))).))..))))))))))))). ( -33.50) >DroSim_CAF1 57741 120 + 1 GCAUUUGUAGCGCCGCAUCUUCUUCUUCUUCCCCUCUUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCGCUGACAAUCGGCGCUGCAAC ....(((((((((((((............(((......))).................(((....)))..))).((((((...((((...))))...))))))......)))))))))). ( -32.30) >DroEre_CAF1 57648 114 + 1 GCAUUUGUAGCGCCGCAUAUU------CUGCCCUUCUUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCGCUGUCAAUCGGCGCUGCAAC ....(((((((((((......------....((.....)).................((((((..(((.(((((....)))))((((...))))...)))..))))))))))))))))). ( -34.50) >DroYak_CAF1 58426 114 + 1 GCAUUUGUAGCGCCGCAUCUU------CUGCCCUUCCAGGAUUAUUUAUUAACGUUUAUUGAUUCCGACUUGCCCGGCGGCAAUCUGCCACAGAAAAUCGCUGUCAAACGGCGCUGCAAC ....(((((((((((....((------(((......))))).................(((((..(((.(((((....)))))((((...))))...)))..))))).))))))))))). ( -36.10) >DroAna_CAF1 48235 111 + 1 GCAUUCGUGGAGCAGCAUCUU---------CCCUUCUUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCUUUGUCAUCCGCUACUGCAAC (((...(((((((((.....(---------((......)))............................(((((....))))).)))).(((((.....)))))..)))))...)))... ( -22.20) >consensus GCAUUUGUAGCGCCGCAUCUU______CUGCCCCUCUUGGAUUAUUUAUUAACGUUUAUUGAUUUCGACUUGCCCAGCGGCAAUCUGCCACAGAAAAUCGCUGUCAAUCGGCGCUGCAAC ....(((((((((((................((.....))..................(((((..(((.(((((....)))))((((...))))...)))..))))).))))))))))). (-26.89 = -27.95 + 1.06)


| Location | 12,675,179 – 12,675,293 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -28.92 |
| Energy contribution | -30.33 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12675179 114 - 22407834 GUUGCAGCGCCGAUUGACAGCGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCACGAGGGGCAG------AAGAUGCGGCGCUACAAAUGC .(((.((((((.(((((...((((((((((((((.....))))).))).))))))...)))))................(((....)))(((.------....))))))))).))).... ( -36.70) >DroSec_CAF1 55853 114 - 1 GUUGCAGCGCCGAUUGUCAGCGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCACGAGGGGCAG------AAGAUGCGGCGCUACAAAUGC .(((.((((((.((((....((((((((((((((.....))))).))).))))))....))))................(((....)))(((.------....))))))))).))).... ( -35.40) >DroSim_CAF1 57741 120 - 1 GUUGCAGCGCCGAUUGUCAGCGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCAAGAGGGGAAGAAGAAGAAGAUGCGGCGCUACAAAUGC .(((.(((((((((((....((((((((((((((.....))))).))).))))))....))))................(((......)))..............))))))).))).... ( -35.80) >DroEre_CAF1 57648 114 - 1 GUUGCAGCGCCGAUUGACAGCGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCAAGAAGGGCAG------AAUAUGCGGCGCUACAAAUGC .(((.((((((.(((((...((((((((((((((.....))))).))).))))))...))))).................((.....))(((.------....))))))))).))).... ( -36.00) >DroYak_CAF1 58426 114 - 1 GUUGCAGCGCCGUUUGACAGCGAUUUUCUGUGGCAGAUUGCCGCCGGGCAAGUCGGAAUCAAUAAACGUUAAUAAAUAAUCCUGGAAGGGCAG------AAGAUGCGGCGCUACAAAUGC .(((.((((((((((((...((((((((((((((.....)))).)))).))))))...))))...................(((......)))------.....)))))))).))).... ( -34.90) >DroAna_CAF1 48235 111 - 1 GUUGCAGUAGCGGAUGACAAAGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCAAGAAGGG---------AAGAUGCUGCUCCACGAAUGC ((.(.(((((((..(((....(((((((((((((.....))))).))).)))))....)))..................(((......))---------)...)))))))).))...... ( -26.20) >consensus GUUGCAGCGCCGAUUGACAGCGAUUUUCUGUGGCAGAUUGCCGCUGGGCAAGUCGAAAUCAAUAAACGUUAAUAAAUAAUCCAAGAAGGGCAG______AAGAUGCGGCGCUACAAAUGC .(((.((((((((((((...((((((((((((((.....))))).))).))))))...)))))................(((.....)))...............))))))).))).... (-28.92 = -30.33 + 1.42)

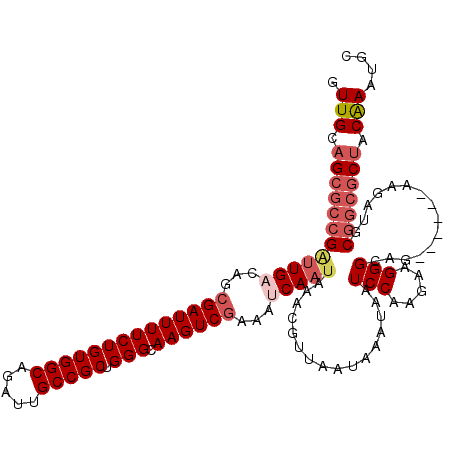

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:06 2006