| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,673,194 – 12,673,300 |
| Length | 106 |
| Max. P | 0.947766 |

| Location | 12,673,194 – 12,673,300 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -16.31 |
| Energy contribution | -15.23 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
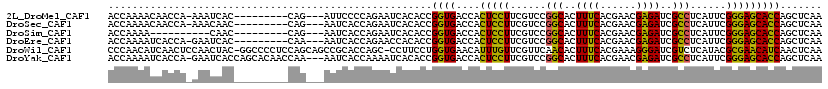
>2L_DroMel_CAF1 12673194 106 + 22407834 ACCAAAACAACCA-AAAUCAC---------CAG---AUUCCCCAGAAUCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA .............-......(---------(.(---((((....))))).....(((((...(((.(((((...........))))).))).)))))......))((((....)))).. ( -24.60) >DroSec_CAF1 53753 106 + 1 ACCAAAACAACCA-AAACAAC---------CAG---AAUCACCAGAAUCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA .............-......(---------(.(---(.((....)).)).....(((((...(((.(((((...........))))).))).)))))......))((((....)))).. ( -21.40) >DroSim_CAF1 55333 97 + 1 ACCAAAA----------CAAC---------CAG---AAUCACCAGAAUCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA .......----------...(---------(.(---(.((....)).)).....(((((...(((.(((((...........))))).))).)))))......))((((....)))).. ( -21.40) >DroEre_CAF1 55527 106 + 1 ACCAAAAUCACCA-GAAUCAC---------CAA---AAUCACCAGAACCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA ..........((.-((((...---------...---..(((((...........)))))..............(((..((((......))))..)))..))))))((((....)))).. ( -20.60) >DroWil_CAF1 6754 117 + 1 CCCAACAUCAACUCCAACUAC-GGCCCCUCCAGCAGCCGCACCAGC-CCUUCCUGGUGAACAUUUGUUCGUUCAACACUUUCACGAAAGGGAUCGUCUCAUACGCGAACAUCAACUCAA ....................(-(((..(....)..))))((((((.-.....))))))......(((((((.....((..((.(....).))..)).......)))))))......... ( -24.00) >DroYak_CAF1 56273 115 + 1 ACCAAAAUCACCA-GAAUCACCAGCACAACCAA---AAUCACCAAAAUCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA ..........((.-((((.....((........---..(((((...........)))))...(((.(((((...........))))).)))...))...))))))((((....)))).. ( -22.00) >consensus ACCAAAAUCACCA_AAACCAC_________CAG___AAUCACCAGAAUCACACCGGUGACCACUCCUUCGUCCGGCACUUUCACGAACGAGAUCGCCUCAUUCGGGAGCACCAGCUCAA ......................................................((((....(((((......(((..((((......))))..)))......)))))))))....... (-16.31 = -15.23 + -1.08)
| Location | 12,673,194 – 12,673,300 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
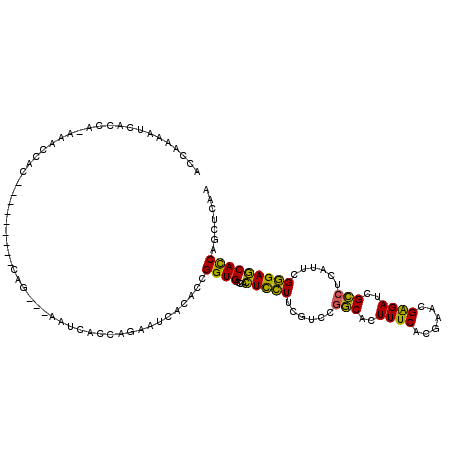
>2L_DroMel_CAF1 12673194 106 - 22407834 UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGAUUCUGGGGAAU---CUG---------GUGAUUU-UGGUUGUUUUGGU .....((((..(((.((((((((.....)))))))).))..)..)))).(((((.((.((((((((....(((((....))))---)))---------)))))).-...)).))))).. ( -35.40) >DroSec_CAF1 53753 106 - 1 UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGAUUCUGGUGAUU---CUG---------GUUGUUU-UGGUUGUUUUGGU ..((((....)))).((((((((.....)))))))).((((..(((((((((.(((...((((((((.......)))))))))---)).---------.))).))-))))..))))... ( -34.90) >DroSim_CAF1 55333 97 - 1 UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGAUUCUGGUGAUU---CUG---------GUUG----------UUUUGGU .....((((..(((.((((((((.....)))))))).))..)..))))((((.(((...((((((((.......)))))))))---)).---------.)))----------)...... ( -34.00) >DroEre_CAF1 55527 106 - 1 UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGGUUCUGGUGAUU---UUG---------GUGAUUC-UGGUGAUUUUGGU .....((((..(((.((((((((.....)))))))).))..)..)))).((..((((((((((((((.......)))))))))---...---------...))))-)..))........ ( -34.80) >DroWil_CAF1 6754 117 - 1 UUGAGUUGAUGUUCGCGUAUGAGACGAUCCCUUUCGUGAAAGUGUUGAACGAACAAAUGUUCACCAGGAAGG-GCUGGUGCGGCUGCUGGAGGGGCC-GUAGUUGGAGUUGAUGUUGGG .....(..(((((.((............((((((((((((..((((.....))))....)))))..))))))-)(..((((((((.(....).))))-))).)..).)).)))))..). ( -35.30) >DroYak_CAF1 56273 115 - 1 UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGAUUUUGGUGAUU---UUGGUUGUGCUGGUGAUUC-UGGUGAUUUUGGU ..((((....)))).((((((((.....)))))))).((((..(((((((....)....((((((((((..(((.........---..)))..))))))))))))-))))..))))... ( -39.40) >consensus UUGAGCUGGUGCUCCCGAAUGAGGCGAUCUCGUUCGUGAAAGUGCCGGACGAAGGAGUGGUCACCGGUGUGAUUCUGGUGAUU___CUG_________GUGAUUC_UGGUGAUUUUGGU ....(((((..(((.((((((((.....)))))))).))..)..)))).).........((((((((.......))))))))..................................... (-26.31 = -25.98 + -0.33)

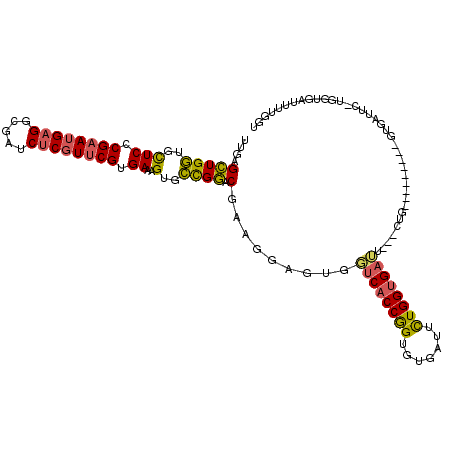
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:01 2006