| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,307,054 – 1,307,158 |
| Length | 104 |
| Max. P | 0.640016 |

| Location | 1,307,054 – 1,307,158 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
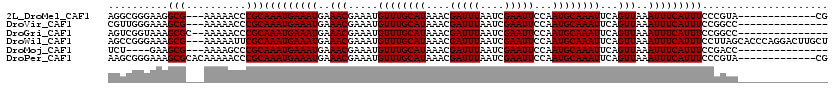
>2L_DroMel_CAF1 1307054 104 - 22407834 AGGCGGGAAGGCG---AAAAACCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCCGUA-------------CG ..(((((((((((---.......)))...((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))))))).-------------.. ( -29.20) >DroVir_CAF1 104852 102 - 1 CGUUGGGAAAGCG---AAAAACCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCGGCC--------------- .((((((((.(((---.......)))...((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))))))).--------------- ( -25.90) >DroGri_CAF1 94348 103 - 1 AGUCGGUAAAGCGC--AAAAACCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCGGCC--------------- .(((((....(((.--.......)))(((((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))))))).--------------- ( -26.10) >DroWil_CAF1 94920 117 - 1 AGCCGGGAAAGCG---AAAAAUUCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCUUAGCACCCAGGACUUGCU ..(((((...(((---(.....))))(((((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))........))).))....... ( -28.10) >DroMoj_CAF1 131780 98 - 1 UCU----GAAGCG---AAAAGCCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCGACC--------------- ...----...((.---....)).((.(((((((((..(((.....((((((((....(((((....)))))...))))))))...)))..))))))))).))...--------------- ( -20.30) >DroPer_CAF1 99221 107 - 1 AAGCGGGAAAGCGCACAAAAACCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCCGUA-------------CG ..(((((((((((..........)))...((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))))))).-------------.. ( -28.20) >consensus AGUCGGGAAAGCG___AAAAACCCGCAAAUGAAAUGAAACGAAAUGUUUGCAUAAACGAUUUAAUCGAAUUCCAAUGCAAAUUCAGUUAAAUUUCAUUUCCGGCC_______________ ..........(((..........)))(((((((((..(((.....((((((((....(((((....)))))...))))))))...)))..)))))))))..................... (-17.90 = -17.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:17 2006